Research Article
Compound of Action in DNA is to be the Internal One Coming from Carbon Value
954
Views & Citations10
Likes & Shares
In order to test our earlier work on carbon value associated with protein structure and function in dealing with biological activities, it is extended here in DNA system where the molecular DNA ought to be binding with drug with the principle arrived at carbon value association. We have calculated the internal one coming from carbon value of association for DNA and its complex with drug. It reveals that the association comes purely from carbon value based on cohesiveness. Accordingly, one can go on applying this carbon value-based force for cohesiveness and development of new drug. This carbon value calculations going to be crucial for drug discovery and development in pharmaceutical industry.
Keywords: Drug-DNA, Carbon domain, Active site, Internal COD, Carbon value
EARLIER WORK
Earlier work on carbon analysis and all worked out with principle that protein structure and association are due to cohesiveness in the internal nature of association [1-4]. Accordingly, the amino acids are organized to meet this internal one [5]. At the same time the thymine centered codon does play an important role in providing adequate hydrophobic residues [6-9]. Arranging these hydrophobic residues are crucial for local structure to be intact [10]. Otherwise going to be inadequate in carbon role which might be attracting incoming molecules to satisfy its carbon value. Based on this carbon score, we have studied the role of carbon in disease [11-13] and applied to several applications [14-17]. According to carbon score, one would expect a carbon value of 31.45% for stable association internally in the macromolecule [18-22]. Accordingly, we have tested this principle of carbon score for protein system and all. Very well understood the concept of binding and associated changes in the system internally and all. After those study, it is extended here in DNA system where drug do interact.
We have been working on for some time now that protein association with other probe one is based on carbon value coming with cohesiveness [23-28]. Accordingly, the association might be playing a role in other system as well including in DNA, RNA, carbohydrate and other carbon biopolymers. One can go on studying these carbon value-based association study for different systems. One such a system here is drug-DNA interaction and all.
Methodology
Data
X-ray crystal structure of DNA (3U2N) and drug-DNA(1DNH) complex are obtained from PDB web located in RCSB. Otherwise, all of the protein and nucleic acid structures determined by various methods are stored here in PDB site of international collaboration and all. We have collected several structures of DNA interest but only those above mentioned are taken for further evaluation. The oxygen atoms of the water molecules are removed from PDB one. There are some Mg atoms in DNA crystal structure which is not there in complex. The Mg atoms are included in the internal COD calculations. Hydrogen atoms are added to the crystal one using online tools available freely for the scientific community.
Draft ICOD
As in protein structure evaluation, the DNA structure also evaluated for internal one using our home-made PERL program. Fantastic observations are noticed here in DNA system of drug binding site and all. Like in protein, it is observed to be the dia16 works out very well for the internal one and all. Interestingly each nucleotide is considered to be double the size of an average amino acids. That is to say the average amino acid size is 15.5555 atoms per residue where are in DNA it is about 32 atoms per residues. So, the deviations in active one considered for proteins is about 77 atoms approximately 5 residues long whereas in DNA it can be 2.5 nucleotides which is about 80 atoms.
The internal COD value for DNA and drug-DNA structures are obtained via ICOD software used in protein evaluation. Same dia16 is used get the ICOD values. After that the points are evaluated using plot as shown in Figure 1.
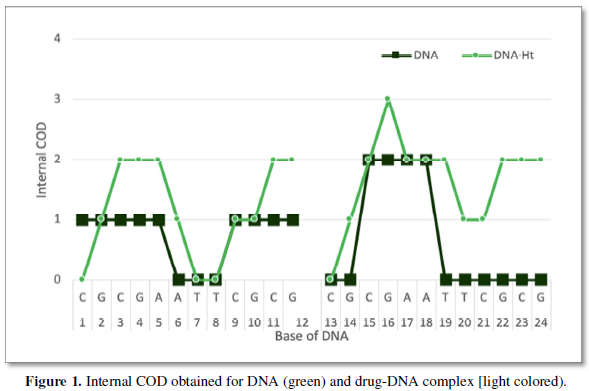

RESULTS AND DISCUSSION
The DNA sequence of ICOD valuation is given in Figure 2. Palindrome of DNA sequence with all of an adenine centered in ICOD of DNA obtained using our home-made software is shown here as model one as XY plot. Figure 1 indicate the ICOD of all obtained values in DNA and drug one against number which are labelled here in. Anything above zero are considered to be ICOD and treated as stable portion. But when it is zero, the ICOD is not followed and unstable portion. We notice here that there are internal COD but 6-8 and 19-24 are long non internal COD regions which are considered to be active site. On binding the drug one this active site, it becomes internal COD and all. That is to say that the active site is non non-COD stretch now. As long the drug is interacting the, there are no COD and stable. The stability of this kind of binding is favored by the cohesiveness coming from carbon value which are governed by formula value of carbon 31.45% here in ICOD calculation and all.


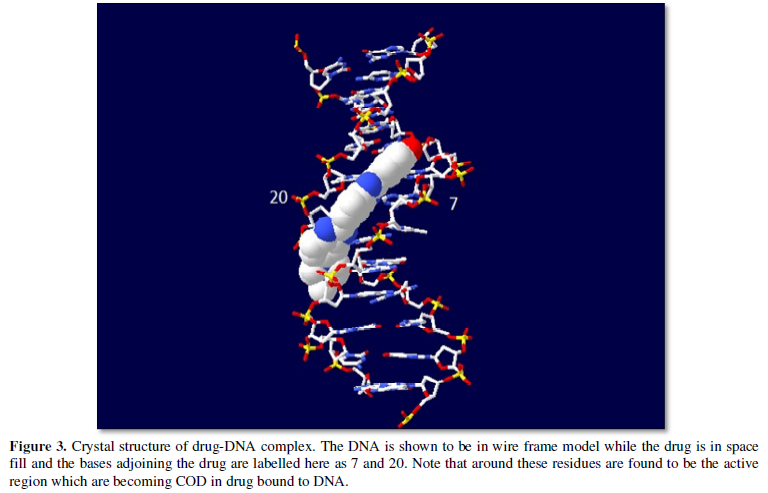
Of course, the ICOD obtained may be altered by changing this drug one with some of other functional one or atoms to adjust parameter of ICOD one. If alteration favors better binding capability, well the drug may be superior over the current one. Accordingly, one can go on synthesizing better drug one for the current existing drugs world. Practically speaking synthesizing totally new drug may be difficult at this point of time. But may be latter with sophisticated technology and all possible to do design. Alternatively, one would go on finding solution at root level that is gene modification and all. Favorable are to be chosen from atomic point of view rather than molecular interaction studies and all. Atomic solution based on carbon value are to be the best solution for finding drug and all (Figure 3).

CONCLUSION
Drug one going to be the apt for all those suffering and all. At this time of research, adequate principles that govern drug discovery are not yet developed. However, the new finding here in conjunction with drug-DNA interaction based on carbon value is promisingly the future of pharmaceutical industry to focus. Carbon value-based finding of drugs going to be the future of drug discovery and all. All that is hopefully going to solve all problems associated with gene malfunction and associated protein interactions. It is only demonstration here that drug can be better worked out at atomic level with accurate binding capability for neat selection for particular site to be focused.
- Rajasekaran E, Meenal R, Indupriya R, Prabakaran TR, Boobalan S (2019) Existence of cohesive force explains all phenomena that are in material which holds strong bond of all forces of attraction: A case study with carbon material., AIP Conference Proceedings 2087, 020015.
- Rajasekaran E, Meenal R, Indupriya R (2020) Internal carbon of chemical molecules play in intermolecular association favoring stability. Materials today: Proceedings.
- Rajasekaran E, Meenal R, Indupriya R (2020) Study on aquaporin proves to be the carbon in protein-protein interface playing in tetramerization. High Technol Lett 26(5): 292-298.
- Rajasekaran E, Indupriya R, Meenal R (2019) Domain formation in regions of protein probe interaction. Int J Mol Biol Opn Acc 4(5): 167-169.
- Rajasekaran E, Kavitha V, Ganesh Bbabu GP, Prabakaran R, Meenal R (2019) Nature of amino acid sequence instruct carbon value to be adopted in protein 3D structure. IEEE Access 1054-1060.
- Rajasekaran E, Indupriya R, Meenal R (2020) Change of corona nucleic acid for human disease suffering. J Xidian Univ 14(3): 1759-1767.
- Balamurugan P, Rajasekaran E, Klaus H (2013) Thymine distribution in genes provides novel insight into the functional significance of the proteome of the malaria parasite Plasmodium falciparum 3D7. Bioinformatics 30(5): 597-600.
- Rajasekaran E, Jacob A, Heese K (2012) Magnitude of thymine in different frames of messenger RNAs. Int J Bioinfo Res 4(3): 273-275.
- Rajasekaran E, Jacob A (2013) Thymine in different frames of untranslated regions of nucleic acid. Int J Bioinfo Res 5(1): 282-284.
- Rajasekaran E, Sheeba K (2013) Carbon distribution accounts a lot for patterns in proteins. Ind J Bioinfo Biotechnol 2(1): 45-47.
- Rajasekaran E, Rajasekaran I (2019) Who power sickle cell disease: Carbon domain analysis tells all because of design in protein 3D arbitrary internal carbon domain (COD) arrangement. Int J Mol Biol Opn Acc 4(3): 85-88.
- Rajasekaran E, John SN, Vennila JJ (2012) Carbon distribution in protein local structure direct superoxide dismutase to disease way. J Proteins Proteomics 3(2): 99-104.
- Akila K, Balamurugan P, Raja E (2012) The nature of proteins in Influenza. Health 4(10): 991-994.
- Rajasekaran E (2013) Carbon distribution in protein structure might influence thermo stability of modified form. J Adv Biotechnol 12(9): 9-10.
- Mamboya FA, Nsimama PD, Amri E, Sharmila JS, Rajasekaran E (2012) Carbon distribution analysis on mutations responsible for Li-Fraumeni syndrome. J Bio Sci 1(2): 1.
- Amri E, Mamboya AF, Nsimama PD, Rajasekaran E (2012) Role of carbon in crystal structures of wild-type and mutated form of dihydrofolatereductase-thymidylate synthase of P. Falciparum. Int J Appl Biol Pharm Technol 3(3): 1-6.
- Nsimama PD, Mamboya AF, Amri E, Rajasekaran E (2012) Correlation between the mutated colour tunings and carbon distributions in luciferase bioluminescence. JCIB Comput Intelli Bioinfo 5(2): 105-112.
- Rajasekaran E (2013) Scale for nature of hydrophobic interactions in proteins. J Proteomics Bioinform 6(7): 31.
- Rajasekaran E, Akila K, Vijayasarathy M, Vinobha CS, Senthil R, et al. (2014) CARd-3D: Carbon distribution in 3D structure program for globular proteins. Bioinformation 10(3): 138-143.
- Rajasekaran E (2012) CARd: Carbon distribution analysis program for protein sequences., Bioinformation 8(11): 508-512.
- Vinobha CS, Rajasekaran E (2010) Atomic details of globular proteins. J Comput Intelli Bioinfo 3(2): 133-136.
- Rajasekaran E, Vinobha CS, Vijayasarathy M, Senthil R Sankarganesh P (2009) The nature of proteins. International association of computer science and information technology, (IACSIT-SC) Singapore 464-465.
- Rajasekaran E, Indupriya R Protein-Ibuprofen interaction: The role of carbon in dealing active site-specific interaction. Adv Nano Mmed Nanotech Res.
- Rajasekaran I, Meenal R, Rajasekaran E (2019) Existence of carbon domain alters bond orders in protein. Int J Innov Eng Technol 13(3): 128-132.
- Indupriya R, Meenal R, Kavitha R, Rajasekaran E (2019) Drug-protein interaction validates the internal COD formed due to cohesive force: Test of bond length variation in amino acids involved. Int J Mol Biol Opn Acc 4(3): 113-117.
- Rajasekaran E (2018) Domains based in carbon dictate here the possible arrangement of all chemistry for biology. Int J Mol Biol Opn Acc 3(5): 240-243.
- Rajasekaran E, Meenal R, Michael PA, Indupriya R (2019) Existence of nano level force in protein plays applications of maximum untold understanding of life form. Int J Eng Adv Technol 9(2): 3722-3726.
- Ekambaram R, Rajasekaran I (2020) Temperature data in 3D structure reveal the internal COD involvement in molecular form. Bioinform Proteom Opn Acc J 4(2): 000136.
QUICK LINKS
- SUBMIT MANUSCRIPT
- RECOMMEND THE JOURNAL
-
SUBSCRIBE FOR ALERTS
RELATED JOURNALS
- Advances in Nanomedicine and Nanotechnology Research (ISSN: 2688-5476)
- Journal of Genetics and Cell Biology (ISSN:2639-3360)
- Journal of Astronomy and Space Research
- Journal of Microbiology and Microbial Infections (ISSN: 2689-7660)
- Food and Nutrition-Current Research (ISSN:2638-1095)
- Journal of Veterinary and Marine Sciences (ISSN: 2689-7830)
- Journal of Biochemistry and Molecular Medicine (ISSN:2641-6948)




