Research Article
Nucleic Acids (DNA/RNA) as Nanoparticles Structures for siRNA Delivery Medical Applications
3552
Views & Citations2552
Likes & Shares
Engineered design of synthetic DNA/RNA molecules can generate pre-defined structures that can easily self-assemble to form nanoparticles with multiple functionalities. The identification and selection of highly potent siRNA sequences has already been accomplished for many gene targets, and the synthesis of siRNAs on a large scale has been achieved. The field of oligonucleotide-based nanotechnology for biomedical applications is just emerging, but will play an important role in the delivery of siRNA. In particular, oligonucleotide-based structural RNAi systems described in this chapter are promising as a new generation of gene delivery carriers for cancer therapy.
RNA interference (RNAi) is a gene regulation mechanism initiated by RNA molecules that enables sequence-specific gene silencing by promoting degradation of specific mRNAs. Molecular therapy using small interfering RNA (siRNA) has shown great therapeutic potential for diseases caused by abnormal gene overexpression or mutation. The major challenges to application of siRNA therapeutics include the stability and effective delivery of siRNA in vivo. In this chapter, we discuss recent advances in nanoparticle-mediated siRNA delivery systems and the application of these systems in clinical trials for cancer therapy. Furthermore, we offer perspectives on future applications of siRNA therapeutics.
Keywords: RNAi, siRNA, Nanotechnology, Structural DNA/RNA
INTRODUCTION
In order to activate the RNAi pathway, double stranded siRNA must travel through the bloodstream and gain access to the cytosol of target cells. The hydrophilic nature and large molecular weight of siRNAs prevent the molecules from diffusing across the cellular membrane into the cell; therefore, modifications to the nucleic acid and generation of clever delivery strategies are necessary for the creation of siRNA therapeutics.
STRUCTURAL DNA/RNA-BASED RNAI SYSTEMS
RNA interference (RNAi) has been recognized as the sequence specific silencing of target mRNA by a long ds-RNA, enabling efficient suppression of gene and protein expression [1]. After the first report on Caenorhabditis elegans in 1998, this phenomenon has been verified in plant, insect, fungi, and mammalian cells [2]. RNAi is now considered to be a highly preserved natural mechanism for the regulation of gene expression in many organisms. Once a long ds-RNA is introduced into the cytoplasm, it is processed by an RNase type-III enzyme (dicer) to generate a short ds-RNA fragment of 21-25 base pairs [3]. The processed ds-RNA fragment can be loaded onto the RNA-induced silencing complex (RISC), and an antisense RNA strand serves as a sequence-specific guide for targeted mRNA cleavage [4,5]. After the dicer process, a short ds-siRNA shows more specific cleavage of target mRNA with improved off-targeting effects.
METHODS
To overcome the critical hurdles of siRNA delivery, various delivery systems have been proposed such as viral and synthetic cationic carrier systems. To date, various viral systems have been developed to endogenously express shRNAs for gene silencing, and these systems include retrovirus, adenovirus, adeno-associated virus (AAV), and lent virus [9]. The viral systems have the clear advantage of a high transduction efficiency and stable expression of shRNAs for a prolonged period. However, many studies have shown the potential drawbacks of viral systems such as risk of mutation, high initial immune response, nonspecific tissue distribution, and undesirable inflammation. Non-viral and synthetic cationic carriers are another class of siRNA delivery system.
Positively-charged polymers, peptides, and lipids have been widely utilized to formulate siRNAs into a compact nanoparticle, facilitating the intracellular uptake of siRNAs [10]. The main mechanism of preparation of nanoparticle complexes is an electrostatic charge interaction between anionic nucleic acids and cationic carriers in aqueous solution. It is similar to that of cationic delivery systems for plasmid DNA (pDNA), however, there is a huge difference in the physical behavior of siRNA compared with long pDNA. siRNA is known to be more rigid due to its rod-like helical structure, having a relatively low charge density, and it remains difficult to formulate compact and stable siRNA complexes with conventional cationic carriers [11,12]. To simply achieve enhanced particle stability, the addition of an excess amount of cationic condensing agent is often carried out to formulate uncompromising siRNAs. However, this non-specific and excessive positive charge on nanocomplexes can cause severe cytotoxicity and immune responses.
RESULTS AND DISCUSSION
Recently, a variety of structural DNA/RNA-based RNAi systems (structural RNAi systems) have been suggested to resolve the aforementioned problems of siRNA delivery. Structural RNAi systems mimic endogenous long ds-RNA, but are prepared by synthetic or equivalent methods to resemble the therapeutic efficacy of siRNAs. The preparation of structural RNAi systems can be as simple as base pair hybridization and bioconjugation, or as complicated as 3D nucleic acid nanotechnology (Table 1). The concept of structural RNAi systems is to overcome the physical drawbacks of siRNAs, while providing structural flexibility to generate more condensed and stable polyelectrolyte complexes. In addition, some of the suggested structural RNAi systems aim to act as carrier-free delivery systems for siRNAs. This approach is particularly unique due to the fact that the structural DNA or RNA itself can serve as a delivery carrier in addition to functioning as a therapeutic drug. In this chapter, we have focused and emphasized the current advances and technological developments in structural RNAi systems. The structural RNAi systems are beginning to show promise, however, the impact on the RNAi field and gene therapy will be realized shortly through the persistent interdisciplinary research in diverse fields.




RNA interference (RNAi) is a process by which RNA molecules, with sequences complementary to a gene’s coding sequence, induce degradation of corresponding messenger RNAs (mRNAs), thus blocking the translation of the mRNA into protein [32,33]. RNAi is initiated by exposing cells to long dsRNA via transfection or endogenous expression. dsRNAs are processed into smaller fragments (usually 21 - 23 nucleotides) of small interfering RNAs (siRNA) [34], which form a complex with the RNA-induced silencing complexes [35]. Introduction of siRNA into mammalian cells leads to downregulation of target genes without triggering interferon responses [34]. Molecular therapy using siRNA has shown great potential for diseases caused by abnormal gene overexpression or mutation, such as various cancers, viral infections, and genetic disorders, as well as for pain management. In the last 10 years, a tremendous effort has been made in biomedical therapeutic application of gene silencing in humans. Phase I studies of siRNA for the treatment of age-related macular degeneration and respiratory syncytial virus provided promising data with no sign of nonspecific toxicity [36,37]. However, there are many challenges to be overcome for siRNA cancer therapeutics, including safety, stability, and effective siRNA delivery.
The major barrier facing siRNA therapeutics is the efficiency of delivery to the desired cell type, tissue, or organ. siRNAs do not readily pass through the cell membrane due to their size and negative charge. Cationic liposome-based strategies are usually used for the cellular delivery of chemically synthesized or in vitro transcribed siRNA [38]. However, there are many problems with lipid-based delivery systems in vivo, such as rapid clearance by the liver and lack of target tissue specificity. Delivery systems can be categorized into physical methods, conjugation methods, and natural carrier (viruses and bacteria) and nonviral carrier methods [39]. DNA-based expression cassettes that express short hairpin RNA (shRNA) are usually delivered to target cells ex vivo by viruses and bacteria, and these modified cells are then reinfused back into the patient [40]. The popular adenovirus- and adeno-associated virus-derived vectors provide efficient delivery for shRNA expression [41]. However, there are problems with delivery using viral vectors, such as insertional mutagenesis and immunogenicity [42]. Nonviral gene delivery systems are highly attractive for gene therapy because they are safer and easier to produce than viral vectors.
Nanotechnology has made significant advances in the development of efficient siRNA delivery systems. Current nonviral delivery systems can be categorized as organic and inorganic [43]. Organic complexes include lipid complexes, conjugated polymers, and cationic polymers, whereas inorganic nanoparticles include magnetic nanoparticles, quantum dots, carbon nanotubes, and gold nanoparticles.
POLY/MULTIMERIC SIRNA DELIVERY APPLICATIONS
In order to properly induce systemic in vivo gene silencing, a large amount of siRNA (3-9 mg/kg) has often been required [44]. However, due to the immune response triggered by excessive RNA materials and cationic carriers, practical applications of RNAi gene therapy have been hampered [45-47]. High molecular weight siRNAs were first proposed in 2007 to improve the physical drawbacks of short rigid ds-RNA [48]. For polyelectrolyte complexation, a more flexible chain of ds-RNA is favorable to form condensed and compact nanoparticles with cationic carriers. However, as compared with plasmid DNA that shows a very flexible nature, siRNA has a rigid rod-like structure with an estimated length of 7 nm. Since the persistent length of ds-RNA is over 260 bp [49-51], siRNA cannot be easily formulated using conventional cationic carriers that are designed for the delivery of pDNA. The physical problems of short ds-RNA have been well-documented in various studies, and the advantages of high molecular weight siRNAs have been highlighted [48,52].
Long linear siRNA
Previously, various methods for preparing long linear siRNA have been investigated. Simple sticky overhang hybridization and bioconjugation of each sense and antisense strand of siRNA has mainly been proposed to generate multimeric blocks of siRNA. Among these, a cleavable disulfide linkage between siRNA strands has been popularly utilized [53-55]. The 5′-ends of the sense and antisense siRNA strand were functionalized with free thiol groups, and these thiol-modified RNA strands were utilized to form a disulfide-polymerized polysiRNA. The polymerized siRNA had a broad range of bp length in the order of 50–1000 bp [13]. In the complexation experiment with a low molecular weight polyethylenimine (LMW-PEI) (MW: 1800), polysiRNA formed condensed and compact nanocomplexes at a weight ratio of 1.25 and a size of 235 nm, while mono- or naked siRNA generated large and loose particles with a size over 1000 nm. The stability of poly-siRNA was also confirmed by a heparin competition assay and serum stability assay. The results revealed that poly-siRNA was far more condensed, overcoming the serum degradation and being more ionically stable than mono-siRNA. In addition, in vitro cellular uptake and gene silencing experiments verified that poly-siRNA has a greater efficiency over mono-siRNA due to its high charge density and stability under formulated conditions.
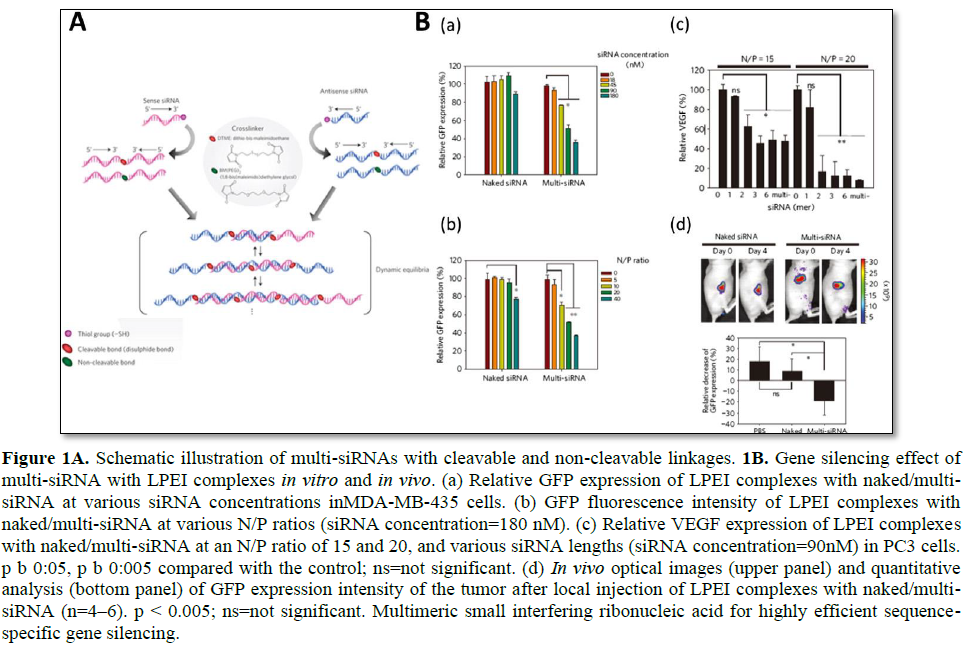
Mok [14] highlighted the in vivo efficacy of long linear siRNA (Figure 1). Multimerized siRNA (multi-siRNA) was prepared by utilizing a cleavable and non-cleavable cross linker [14]. The 3′-end of each RNA strand had a thiol functional group, which later reacted with a short cross linker to produce a multi-siRNA. Since this study utilized both a cleavable and non-cleavable linker, a detailed study of the gene silencing mechanism of multi-siRNA was accomplished. For multi-siRNA with a cleavable linker, once it is internalized, it can dissociate to mono-siRNA due to the reducing conditions in the cytoplasm as a result of glutathione (GSH). However, in the case of multi-siRNA with a non-cleavable linker, dicer is needed to process the multi-siRNA to generate short ds-RNAs by random cleavage. Therefore, the cleavable linker provided a more sequence-specific degradation of mRNA. In vivo experiments have revealed that multi-siRNA show far enhanced gene silencing efficacy as compared with naked siRNA. Immunostimulation upon the injection of multi-siRNA has also been investigated, and both cleavable and non-cleavable multi-siRNA showed a relatively low level of interferon alpha (IFN-α) when formulated with linear PEI (LPEI). It is important to note that, when non-cleavable multi-siRNA was formulated with N- [1-(2,3-dioleoyloxy) propyl]-N, N, N-trimethylammonium methylsulfate (DOTAP), a massive increase in IFN-α was observed. This suggests that immune stimulation is highly affected by not only the genetic material itself but also by the delivery carriers.
Dual gene targeted multimeric siRNA conjugates (DGT multi-siRNA) was developed to induce simultaneous gene knockdown of two selective proteins (green fluorescent protein (GFP) and vascular endothelial growth factor (VEGF)). Using either cleavable or non-cleavable crosslinkers, an anti-GFP and anti-VEGF sequence containing multimeric siRNA was prepared by the thiol-maleimide reaction of the 3′-end of thiol functional group of

the RNA and crosslinker [15]. It is interesting to note that, at the same concentration, the DGT multi-siRNA induced enhanced gene silencing of target proteins as compared with the mixture of single gene-targeted multimeric siRNA. Since simultaneous silencing of multiple upregulated genes is highly attractive for anticancer treatment, the surviving and Bcl-2 genes have been dual targeted, and a synergistic apoptotic effect on cancer cells was achieved. To further evaluate the enhanced gene silencing on dual-targeted siRNA delivery, dimerized siRNA was synthesized with a cleavable disulfide bond [56]. Unlike multi-siRNA that is the mixture of various long ds-RNAs such as a mixed population of multimers, the dimerized siRNA can offer a better-quality control for synthesis. As compared with monomer siRNA, dimerized siRNA showed far enhanced complexation behaviors with cationic polymers. In addition, enhanced intracellular delivery and gene silencing could be achieved with the dimerized siRNA with a polyethylene glycol (PEG) modification to improve the serum stability of nanocomplexes.
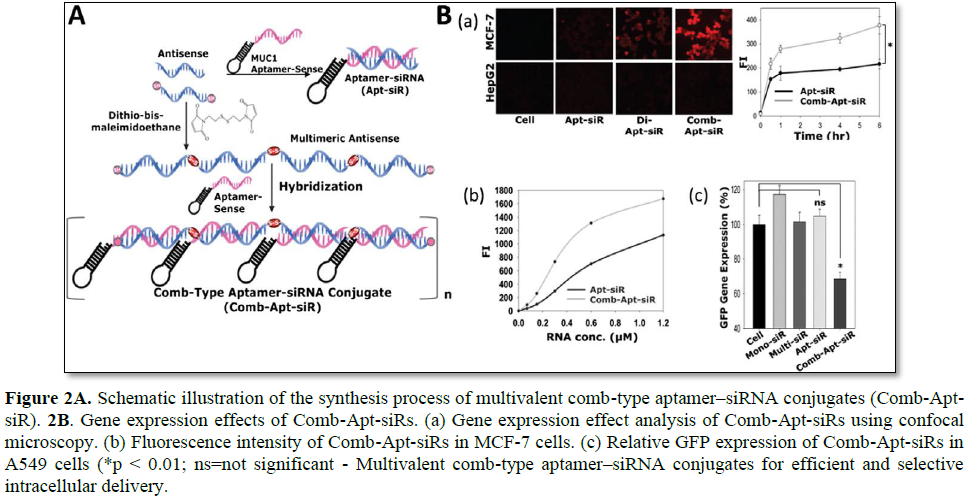
Multimerized siRNA systems have been utilized as a scaffold carrier for the delivery of aptamer-siRNA conjugates (Figure 2). An aptamer is a short oligonucleotide with a high binding affinity for various target molecules including small molecules, peptides, and oligonucleotides. Aptamers have been widely used as novel ligands for targeted gene delivery due to their various advantages such a high specificity and binding affinity, low immune stimulation, and the ease of preparation [18]. Amucin 1 (MUC1) DNA aptamer was used as a targeting ligand for cancer cells becauseMUC1 is highly overexpressed in malignant adenocarcinoma [10,18]. To prepare the comb-type aptamer–siRNA conjugates, antisense strands of siRNA were first multimerized with a cleavable disulfide linkage, and later MUC1 aptamer-sense strands were hybridized to form a linear multimerized dsRNA structure with repeated introduction of MUC1 aptamers. As compared with the direct one to one conjugate of MUC1 aptamer and siRNA, enhanced uptake of the comb-type aptamer–siRNA conjugates were achieved in MCF-7 cells. It is likely that this enhanced intracellular uptake of multivalent comb-type aptamer-siRNA conjugates (Comb-Apt-siR) is attributed to a more favorable chance of contact with MUC1 and/or the synergistic effects of multivalent aptamers for endocytosis. However, gene silencing of Comb-Apt-siR without an additional cationic carrier was not achieved due to the lack of endosomal escaping properties. This study highlighted the multivalent ligands can greatly enhance the intracellular delivery efficiency of Comb-Apt-siR [19].


Branched siRNA
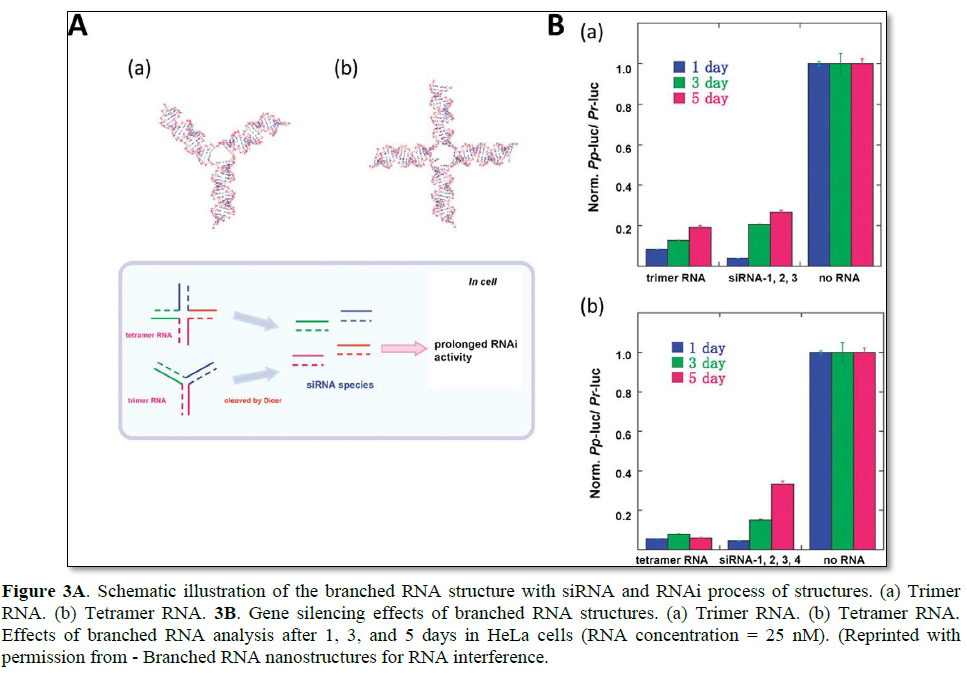
As an alternative to long linear siRNA, branched and dendrimer-like siRNA structures have been developed. Unlike the natural RNA strands that are quickly degraded in biological fluid, functional RNA structures may provide a prolonged RNAi effect due to their increased serum stability and charge density [57]. Nakashima et al. reported the synthesis of a branched siRNA structure with three- or four-way junctions [16]. By simple base pair hybridization, trimer and tetramer RNA were efficiently self-assembled (Figure 3). To evaluate their prolonged RNAi effects, these branched RNA structures were incubated with the dicer, and their stability against nuclease was investigated. Compared with the linear RNA substrate, the branched RNA structures produced active 21 bp siRNAs at a much slower rate. The prolonged generation of siRNAs under cytosolic conditions can enhance the overall duration of the RNAi effect, thereby maximizing gene regulation under various applications. The tetramer RNA resulted in stable luciferase gene silencing over a period of 5 days without chemical modification of RNA bases.
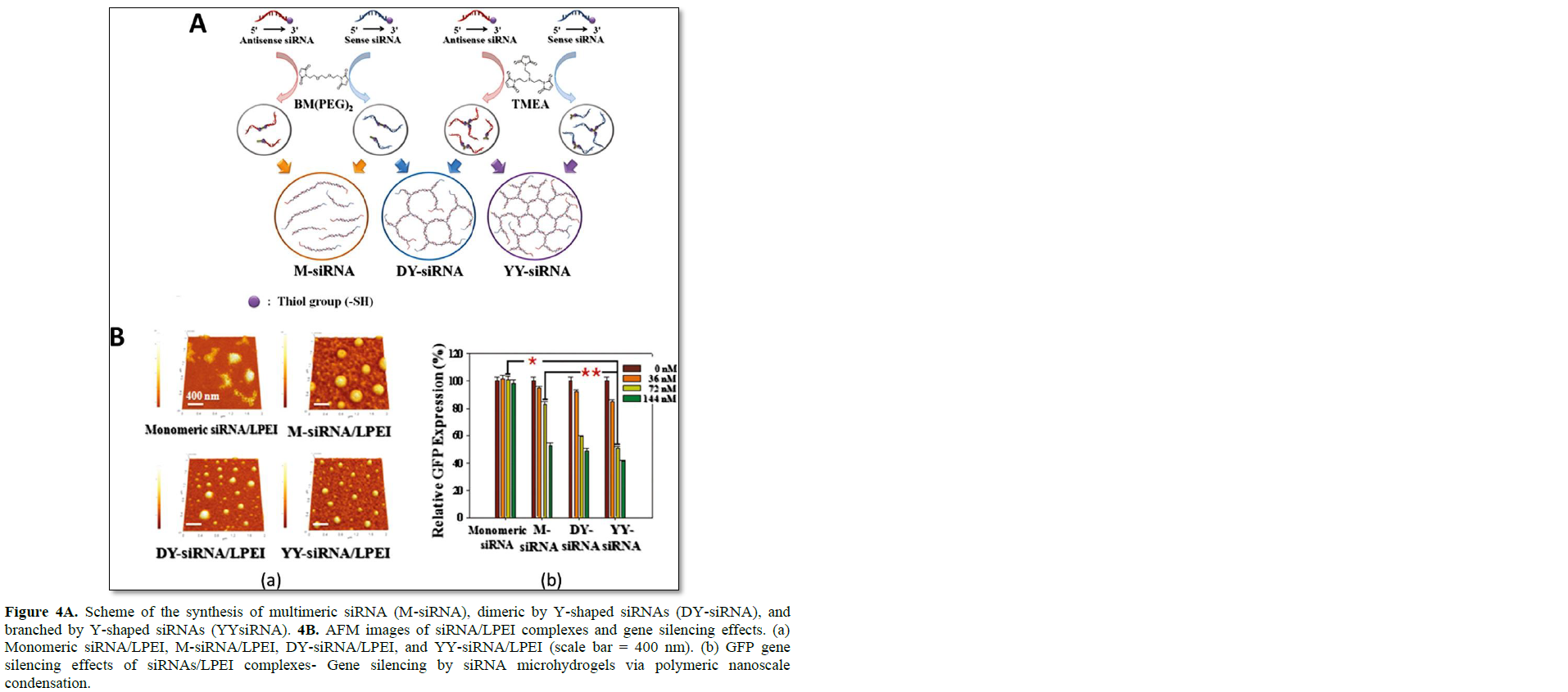
In addition to the simple branched RNA structure, more complex dendrimer-like structures of siRNAs were developed by Hong [17]. Two types of RNA dendrimer were prepared by the simple hybridization of dimeric siRNA and Y-shaped siRNA (Figure 4). The dimeric and Y-shaped siRNAs were synthesized by the use of a non-cleavable crosslinker of 1,8-bis(maleimidodiethylene) glycol (BM(PEG)2) and tri-[2-maleimidoethyl]-amine (TMEA). Depending on the mixture of dimeric and Y-shaped siRNA, highly branched and dendrimer-like structures were generated, and this networked structure could form RNA micro hydrogel. The porosity and networked structure of RNA micro hydrogel could be controlled by simply increasing the ratio of Y shaped siRNA over dimeric RNA. The size of RNA micro hydrogel is roughly 2 μm in the dried state and 8 μm in the well-swollen state in aqueous solution. The highly networked structure of siRNA micro hydrogel offers a greater charge density that facilitates the complexation with a weakly-charged cationic carrier such as LPEI (MW: 2500). At a nitrogen/phosphate (N/P) ratio of 60, highly condensed nanocomplexes could be prepared with a size of 120 nm. The compact nanocomplexes of RNA hydrogel and LPEI under 150 nm in size were highly efficient for inducing the internalization of these particles into cells via an endocytic pathway. In addition, due to more favorable condensation with mild cationic polymers, the siRNA micro hydrogel exhibited far enhanced gene silencing compared with the monomeric and multimerized siRNA, with negligible cytotoxicity. At a siRNA concentration of 72 nM, the siRNA micro hydrogel/LPEI complexes showed significant gene silencing effects (52.8% reduction in GFP expression) over the monomeric siRNA/LPEI (37.7% reduction in GFP expression). Dicer processing of a highly networked structure of RNA has been verified. Random ds-siRNA fragments could be generated from siRNA hydrogel, and the processed short ds-siRNA participated in the RNAi mechanism.


Gold nanoparticles (AuNPs) can also be used to prepare multimerized and highly branched RNA structures [58]. AuNPs have received much attention as an excellent nanoplatform for biomolecule conjugation due to their excellent biocompatibility, controllable morphology, and ease of surface functionalization [1–4,58]. Since AuNPs can readily react with thiol-containing RNA molecules, siRNA-immobilized AuNPs have been investigated for efficient cellular uptake and gene inhibition [59–62]. Kong and coworkers utilized 5 nm AuNPs as a platform to build multimeric/branched RNA structures. The 3′-ends of thiol functionalized RNA strands were immobilized to prepare the sense and antisense-AuNPs. Once the two different AuNPs were mixed together, they formed multimerized siRNA (M-siRNA) crosslinked by AuNPs. Due to the distinct optical properties of AuNPs, formation of M-siRNA can be verified by UV-VIS measurement, exhibiting a blue shift in the absorbance spectrum. Under reducing conditions, thiol-modified siRNAs were released from the surface of AuNPs ready for the formation of RISC without dicer processing in the cytoplasm. The prepared M-siRNA by AuNPs with LPEI (25 kDa) showed enhanced intracellular uptake of nanocomplexes as measured by computed tomography (CT) imaging, and efficient targeted GFP and VEGF gene silencing was achieved in MDA-MB-435 cells.
Novel carriers for poly/multimeric siRNA delivery
Various cationic carriers for poly/multimeric siRNA have been developed. Unlike monomeric siRNA that requires a high cationic charge density for stable polyelectrolyte complexation, lower cationically-charged and relatively small molecular weight carriers have been utilized for the delivery of poly/multimeric siRNA [14]. Due to reduced cationic charges on carriers, the serum stability and cytotoxicity of siRNA/carrier complexes have been resolved with more potent gene silencing as compared with that of monomeric siRNA delivery [63,64]. For instance, thiolated glycol chitosan (TGC) polymer has been developed to formulate a stable nanoparticle structure with poly-siRNA through charge–charge interaction and chemical crosslinking [65]. Upon weak-charge interaction with poly-siRNA, the TGC first formed loosely bound structures, enabling the tight crosslinking of glycol chitosan (GC) polymers for the generation of more condensed nanostructures (size ~300 nm). The condensed nanocomplexes were sensitive to reducing conditions, and 10 mM dithiothreitol (DTT) treatment allowed the total dissociation of monomeric siRNA from the complexes. There were several advantages of TGC polymers as compared with strong cationic polymers such as PEI. When TGC polymers formed nanocomplexes with polysiRNA, their surface was slightly


positive (zeta potential of 3.55 mV) and showed enhanced stability against physiological anionic proteins or carbohydrates. This allowed the effective passive targeting of tumors by systemic injection of poly-siRNA/TGC complexes. Unlike the strong cationic PEI carriers that tend to accumulate in the liver, the TGC carriers exhibited reduced non-specific accumulation in the liver and other organs, and extremely high accumulation in the tumor tissue. The therapeutic efficacy of poly-siRNA/TGC has been tested in tumor-bearing mice, and effective tumor suppression has been achieved upon systemic injection of anti-VEGF poly-siRNA.
Transferrin (TF), a serum protein, is considered to be a good candidate for an efficient siRNA carrier due to its biocompatibility and tumor targeting ability. Since transferrin receptors (TFR) are highly overexpressed in many types of cancer cells, the active targeting of TFR and the intracellular delivery of therapeutics to tumor tissue have been attempted [66]. However, natural TF does not have sufficient binding affinity for the short nucleic acid drugs such as siRNA, thus is unable to serve as a carrier for genetic drugs. To improve the molecular interaction between RNA drugs and TF, poly-siRNA has been utilized along with thiolated TF (tTF). Due to the increase in charge interaction of polysiRNA and tTF, poly-siRNA/tTF could generate a loosely conjugated state followed by a tight condensation process to form nanoparticles through the disulfide crosslinking of tTF. To prepare tTF, TF was functionalized with 2-iminothiolanes under oxygen-limiting conditions, and approximately 8.7 free thiol groups were introduced on TF. Poly-siRNA and tTF formed stable complexes with a mixing ratio of 1:10 (w/w). The particle size before and after the crosslinking process was measured by dynamic light scattering (DLS), and the size of complexes was decreased to 343 nm. The formulated poly-siRNA/tTF complexes were systemically introduced to tumor bearing mice and their biodistribution was obtained by real-time in vivo near-infrared fluorescence (NIRF) imaging. The results confirmed that siRNA/tTF complexes showed tumor-specific active targeting as well as moderate accumulation of these particles in the liver, spleen, and kidney. Since TF served as both an active targeting ligand and a carrier, there was no use of additional cationic materials in this study of this chapter, and effective target-gene silencing in vitro and in vivo was achieved without cytotoxicity.
Similar to transferrin, human serum albumin (HSA) has been widely utilized as a drug carrier, owing to its excellent physical and biological properties such as water solubility, plasma stability, low toxicity, and reduced immunogenicity [67,68]. It is also known that HSA shows a relatively high uptake in tumor and inflamed tissue. Abraxane is the best example of a commercialized albumin-based formulation for anticancer drug delivery, and under cellular stress-inducing conditions, HAS been preferentially taken up by fast-growing tumors as the main energy source for growth and maintenance. For successful encapsulation of poly-siRNA within HSA, the thiolated HSA (tHSA) was synthesized by reacting the amine groups of albumins with Traut's reagent [69]. Thiolated HSA was utilized to encapsulate the poly-siRNA by self-cross linking via disulfide bond formation, generating stable poly-siRNA/HSA nanoparticles. The optimized formulation resulted with nanoparticles under a size of 200 nm, which showed enhanced cellular uptake by albumin transcytosis, and effective in vitro gene silencing was achieved at a siRNA concentration as low as 50 nM. In vivo systemic gene silencing testing has revealed tumor-specific accumulation of poly-siRNA/tHSA complexes and the induction of effective tumor suppression over a period of 30 days. Therefore, along with poly-siRNA, the self-crosslinked HSA nanocarrier system could be a potential candidate for the systemic delivery of siRNA therapeutics for safe and effective anticancer gene therapy.
Three-dimensional RNA/DNA structures for siRNA delivery applications
Although the conventional formulation of siRNA delivery using cationic materials has shown some promise in in vivo animal studies, non-specific charge interaction-driven complexation of cationic carriers and anionic siRNAs have shown multiple drawbacks. These include the heterogeneous size, composition, and surface chemistry of the formulation. Due to the lack of precise control of such properties, varied in vivo biodistribution and pharmacokinetics have been observed, as well as a lack of correlation between in vitro and in vivo studies. Consequently, undesirable and unpredictable in vivo performance of cationic carriers has been reported elsewhere [2,64,70]. To overcome the current problems of cationic delivery carriers, various self-assembled structures of short oligonucleotides have been explored. DNA and RNA molecules are genetic and informative materials; however, they can also serve as an excellent genuine material to prepare more complex and higher ordered structures for various applications [71]. As a result of the good biocompatibility and biodegradability of DNA and RNA molecules, multifunctional self-assembled nanoparticles were prepared for drug delivery by a simple programmable hybridization of complementary strands. These nucleic acid nanoparticles clearly show structural and compositional advantages over the conventional carriers, and various siRNA delivery applications are highlighted in this chapter.
RNA-based nanoparticles for siRNA delivery
Various naturally-occurring RNA structures have been utilized in living cells as gene regulatory materials for mRNA transcription, maturation, translation, degradation, and the catalytic activity of ribozymes [72,73]. For synthetic RNA-based nanoparticles, multiple advantages can be attained as a nanocarrier such as a compact and defined size for tumor targeting, multivalent characteristics for various conjugation, and ease of chemical modification. Naturally- or synthetically generated RNA motifs and modulus can be utilized for assembling nanoparticles with various structural diversities [74-78]. This is particularly attractive to biomedical and clinical applications, since the in vivo fate of nanoparticles is governed by their size, structure, and composition [21,23,24,79-87]. The self-assembled nucleic acid nanoparticles can offer modulation of the therapeutic half-life, biodistribution, cell-specific internalization, and excretion [88]. Moreover, high affinity aptamers can be incorporated into the nucleic acid structures for targeted delivery, and the intracellular uptake process can be more precisely controlled [89].
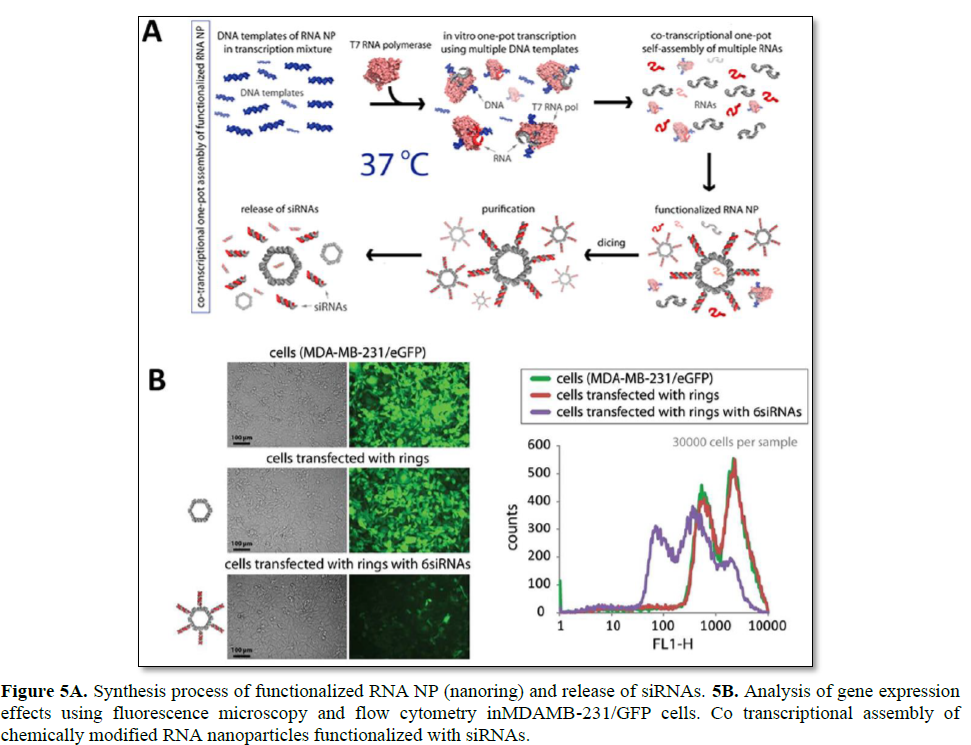
Among various RNA nanoparticles, self-assembling RNA nanoring’s by inverse kissing loop complexation have been reported for the delivery of siRNA (Figure 5) [20]. Among the polygons, a hexamer nanoring structure was selected to provide a thermodynamically stable structure at a relatively low RNA concentration. Assembled nanoring’s have a distinct size and shape that can be later loaded with six siRNAs by simple hybridization on helical stems. Interestingly, the assembled nanoring structures show improved serum stability, but are also processed by dicer to release the loaded siRNAs. When RNA kissing loop complexes with siRNAs were delivered toMDA-MB-231 cells, efficient GFP gene silencing was achieved [22]. Although there exist various advantages of functional RNA nanoparticles, there remain three major drawbacks for siRNA delivery: 1) cost of long synthetic RNA, 2) complexity and difficulty in generating 3D RNA NPs, and 3) short blood half-life due to serum nuclease and renal filtration. To resolve the aforementioned matter, RNA NPs is enzymatically prepared by an in vitro transcription process using T7 RNA polymerase. To optimize the self-assembly conditions, Mn2+ was additionally applied to the incubation solution to result in a high yield preparation of RNA NPs. Lastly, 2′-F-dUMPs can be utilized to generate a 2′-F modification on RNA strands in order to highly improve the serum stability against nucleases [22].


To further apply RNA nanoring’s, multifunctional RNA nanoparticles have been developed for broad applications in nanomedicine. Double stranded RNA is utilized to generate a nanoring scaffold by the toehold interaction. The nanoring scaffold can be further functionalized with various molecules such as siRNAs, aptamers, fluorescence dyes, and proteins. The multifunctional RNA NPs shows enhanced uptake and gene silencing as compared with an equal concentration of duplex RNA molecules. These results are similar to the report of multimeric-siRNA delivery, and prolonged gene silencing was also achieved using the multifunctional RNA NPs. It is likely that the multivalency of RNA nanoring’s can be attributed to the enhanced and prolonged effect of RNAi in vitro. Similarly, for in vivo tumor suppression experiments, the intratumoral injection of multifunctional RNA NPs was conducted, achieving up to a sixfold enhanced GFP gene silencing as compared with ds-siRNA. Finally, functional nanoring’s have been prepared against HIV-1, and their site-specific cleavage against six different regions of HIV-1 has been verified. Complete viral inhibition was obtained at 1 nM concentrations of nanoring’s. Another bottom-up approach of RNA nanotechnology, to generate functional nanoparticles for siRNA delivery, is the use of the DNA- packaging motor of the bacterial virus phi29 [25]. Six copies of packaging RNAs (pRNA) molecules can form a hexametric ring as a critical part of the motor. These molecules can be utilized as a building block to form supramolecular structures such as RNA twins, tetramers, and arrays by the intermolecular self-assembly of palindromic sequences at the 3′- ends of the left and right loop [90–92]. It is known that these self-assembled RNA nanostructures show good stability against changes in temperature, salt, and pH [25]. Due to the small size of these pRNA nanostructures, they have advantages in cell surface interaction as well as internalization. This physical property is particularly pertinent to the delivery of therapeutics and molecular imaging agents, and the bottom-up assembly of pRNA has been applied to the preparation of appropriate delivery scaffolds for such applications.
The trimer pRNA structure, harboring siRNA or other therapeutic molecules, has been fabricated through the interaction of engineered right and left interlocking RNA loops [26]. pRNA with a single strand stem loop does not require additional linkers to incorporate cargo materials within the structure [93]. In fact, the trimer pRNA is multifunctionalized by incorporating aptamers for cell surface targeting, heavy metals, fluorescence dyes, and radioisotopes for molecular imaging and diagnostics [26]. The pRNA/aptamer (CD4) has shown enhanced binding and accumulation of these chimeras on a CD4-overexpressing thymic T cell line. The intracellular entry of the pRNA/aptamer has been confirmed as endocytosis of membrane-bound molecules. Effective in vitro gene regulation by anti-CD4 siRNA has also been achieved in a targeted manner.
Although 117 nt long pRNA has shown effective intracellular delivery of various therapeutics into specific cancer or viral-infected cells, the chemical synthesis of such long RNA is not commercially feasible. To overcome the synthetic drawback, Yi [94] developed a bipartite approach to prepare a pRNA structure from two synthetic RNA fragments with variable modification. The two individual synthetic RNA strands are self-assembled, and readily formed a dimeric pRNA structure similar to that of wild-type pRNA. The viral assembly and DNA packaging activity of bipartite pRNA were compared with wild-type pRNA, and their ability was similar. In addition, an in vitro gene silencing test confirmed that the bipartite pRNA/siRNA could induce effective gene silencing in a targeted manner, with the incorporation of a folate ligand. To improve the multivalency of pRNA, a pRNA-X motif was developed by opening the right-hand loop and introducing a further nine nucleotides within the system [24]. An X motif provides functional arms for four guest molecules, and is prepared by assembling four different RNA oligonucleotide strands. The pRNA-X motif is thermodynamically stable at ambient temperature and can provide multimodule functionalities to carry four different cargo materials such as aptamers, targeting ligands, and siRNA.
DNA polyhedron nanoparticles for siRNA delivery
Various polyhedron DNA nanostructures have been developed by Turberfield [95] and Mao [96]. These include tetrahedron, cube, dodecahedron, and buckyball structures. Similar to that of RNA nanostructures, 3D DNA nanostructures have been explored for imaging and delivery applications [97-99]. Due to the programmable assembly of DNA strands, the size and shape of nanoparticles can be easily controlled. In addition, various polyhedron particles allow precise control of spatial orientation and density of targeting ligands, which cannot be controlled by any other synthetic nanoparticle system.
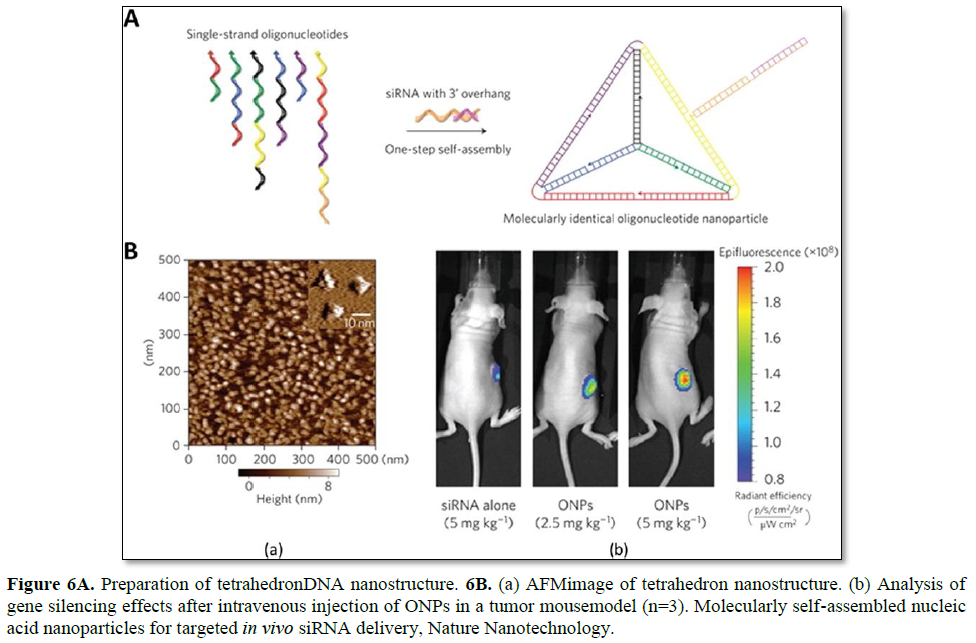
Lee et al. have reported the use of tetrahedron DNA nanoparticles for the targeted in vivo delivery of folic acid-conjugated siRNA (Figure 6). The DNA tetrahedron, consisting of 186 bp, self-assembled from six DNA strands to prepare molecularly-identical oligonucleotide nanoparticles (ONPs). The six edges of the tetrahedron are 30 bp long, with an estimated size of 10 nm. Each edge contains a nick in the middle, where each end of the 5′- and 3′- oligonucleotides meets. To properly incorporate siRNAs into this system, 21-bp overhangs are added to the 3′-end of each DNA strand. As a result, six siRNA strands can be applied to each tetrahedron DNA nanoparticle (one per edge). In addition, for in vivo applications, RNA strands are chemically modified with 2′-OMe to reduce the potential immune response as well as to improve the serum stability [100].
The resultant ONPs have a hydrodynamic diameter of ~28.6 nm, which is suitable for the avoidance of renal filtration, while passively being delivered to tumors via the enhanced permeability and retention (EPR) effect. Since the hybrid DNA/RNA nanoparticles show strong negative surface charges, the intracellular uptake of these particles is not favorable by charge-repulsion between the particles and cell membrane. To enhance the interfacial interaction, various targeting and cell-penetrating ligands have been introduced to the ONP system to facilitate the intracellular uptake of the particles. Targeting ligands are selected from broad materials such as peptides, small molecules, and sugars. Many cationic peptides show false positive enhanced uptake data due to their non-specific interaction with ONPs through electrostatic interactions, forming larger aggregates. Very few cationic peptides allowed particle stability at neutral pH, however these did not show enhanced uptake of the ONPs. Among the screened ligands, folic acid (FA) has shown a concentration-dependent gene silencing effect. FA receptor-overexpressing KB cells were utilized to verify the receptor-mediated uptake of these particles. Since polyhedron ONPs can precisely control the ligand density and location, structure function studies of various ligand densities and orientations on ONPs have been conducted. It has been shown that at least three FA ligands are required to induce appropriate GFP gene silencing, and that their ligand orientation also affects the gene silencing efficiency. It is likely that a higher local FA ligand density may influence the intracellular trafficking pathway of ONPs and the corresponding gene silencing [27].
Large-scale preparation of DNA nanostructures for translational study
There has been much interest in utilizing oligonucleotide-based drug carriers for gene delivery. Despite the advantages

of precise size control, superior intracellular delivery efficiency, and good biocompatibility and biodegradability, many oligonucleotide nanostructures require multiple long nucleotide strands for self-assembly [17]. The cost and synthetic problems in preparing long nucleotides clearly hamper the practical use of DNA or RNA-based nanoparticles. In addition to the synthetic method of preparing long oligonucleotides, there is an enzymatic approach to prepare theses strands that self-assemble to generate DNA nanostructures [101]. Rolling circle amplification (RCA) is one of the examples of enzymatic preparation of DNA nanostructures. This method is a robust technique to generate elongated single-stranded (ss) DNA around a circular ss DNA template under isothermal conditions [102-104].
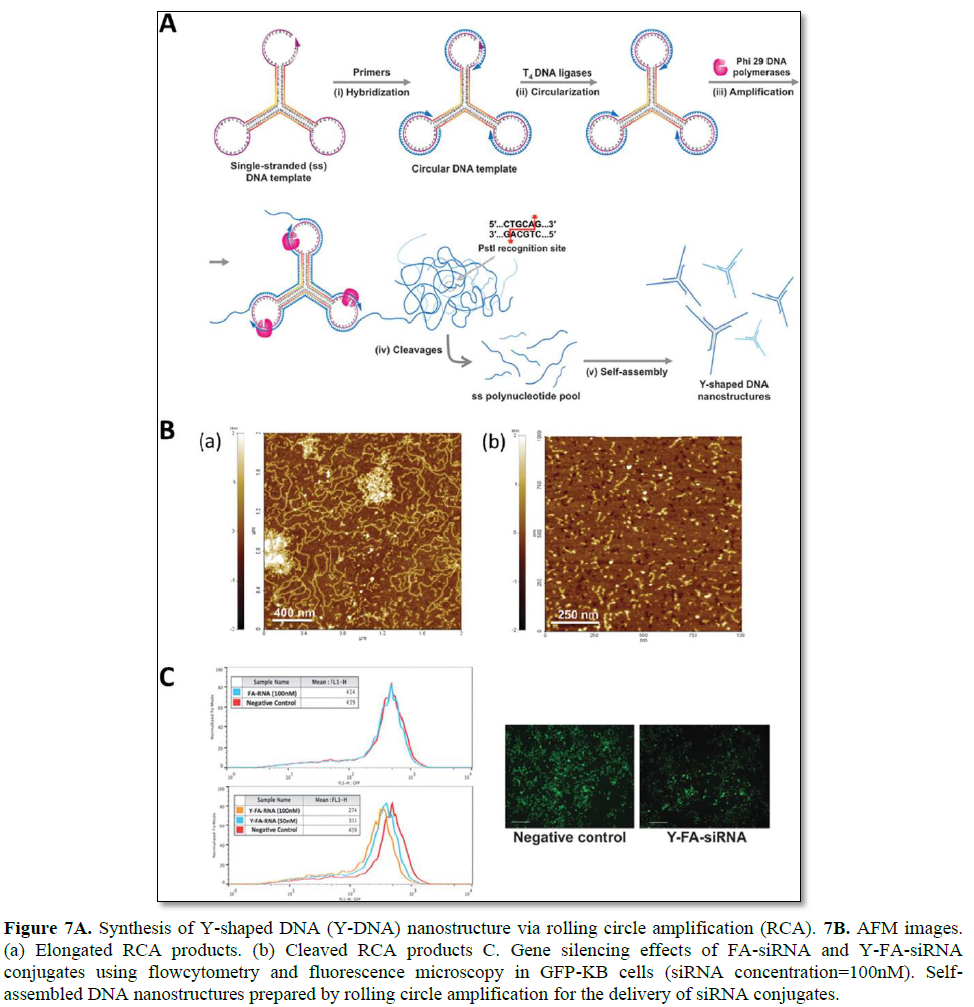
Hong [105] demonstrated RCA-based enzymatic amplification of DNA nanostructures for the delivery of siRNAs (Figure 7). A self-assembled Y shaped DNA structure (159 nt) was used as the closed circular template for RCA to generate long amplified DNA products. Site-specific cleavage can be achieved by containing PstI endonuclease-specific sequences in the open loop of the Y-DNA junction. Inter- and intramolecular self-assembly of an elongated ss DNA product can form hybridization of palindromic PstI sites without the addition of helper DNA strands. After treatment with PstI enzymes, the long ss DNA product is site-specifically cleaved to generate individual DNA fragments, which can later self-assemble to form Y-DNA nanostructures. The overhang sequences on each arm have been designed to form stable hybridization with folic acid-conjugated siRNAs. In this study, 1 pmol DNA template was amplified to approximately 1068 pmol elongated ss DNA, and 213 pmol of Y-DNA structure was produced. This approach is widely useful as a simple platform for the largescale synthesis of various DNA nanostructures for therapeutic applications [105].
DNA/RNA ball technology for siRNA delivery applications
Rolling circle replication (RCR) has been extensively explored to overcome the instability of RNA and the low packing efficiency of the carriers. While conventional polymerase chain reaction (PCR) requires a thermal cycling process and is limited to amplification of short DNA segments, RCR is an isothermal process and is used for the exponential synthesis of long concatemeric DNA/RNA strands by a processive rolling mechanism [106,107]. Rolling circle mechanisms have been widely adapted to different areas including genomics [108], proteomics [109], biosensing [110,111], drug delivery [112,113], and structure building [114,115]. The RCR technique used for gene delivery applications is of special interest. Specifically,


rolling circle transcription (RCT) involves T7 RNA polymerase that can continuously generate multiple single-stranded RNA copies from the template circular DNA. A number of studies have shown that RCT can be used for continued RNA synthesis with high efficiency [107,116,117].
RNA microsponge/ball technology for siRNA delivery
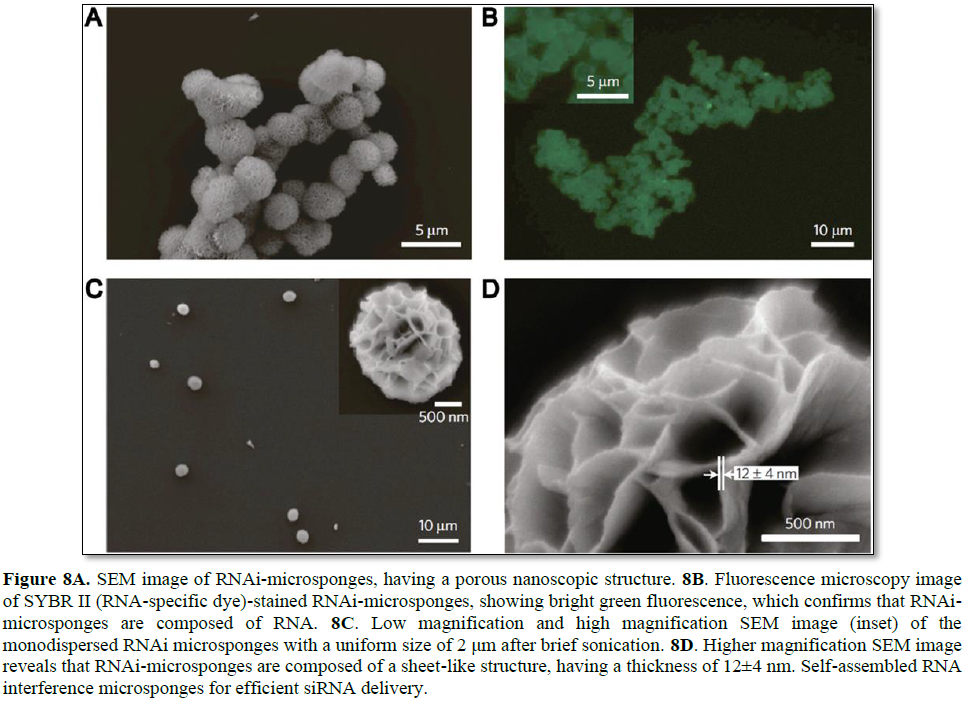
By taking advantage of enzymatic RNA polymerization, condensed RNA structures with predetermined sequences for RNA interference were synthesized [113]. According to the formation process of RNA structures, long replicated RNA strands show similar behavior to traditional synthetic polymers, which is a key factor for efficient siRNA delivery and high cargo capacity. In the early stage of polymerization, a fiber like structure is gradually entangled and forms a sheet-like structure. As these structures grow larger, RNA strands eventually self-assemble into sponge-like spherical structures called RNAi-microsponges (Figure 8). Due to the fact that RNAi-microsponges consist of


multiple copies of tandem RNA units, a large amount of siRNA can be loaded onto a single microsponge. Moreover, it is possible that polyethylenimine (PEI), as a polymeric transfection agent, can be used to condense the microsponge for enhancement of cellular uptake of the particle. Indeed, the RNAi-microsponge/PEI complex showed reasonable silencing efficiency under in vitro and in vivo conditions.
This study demonstrated a new platform for the synthesis of self-assembled RNA structures. By using polymeric RNA strands, spongelike spherical RNA structures can be synthesized, which can achieve a high loading efficiency. Furthermore, RNAi-microsponges which consist entirely of RNA strands are able to rapidly deliver a large amount of siRNA to target cells by simply coating with positively-charged polyions. This novel approach can reduce the limitations of siRNA therapeutics and also be applied to the synthesis of various RNA structures.
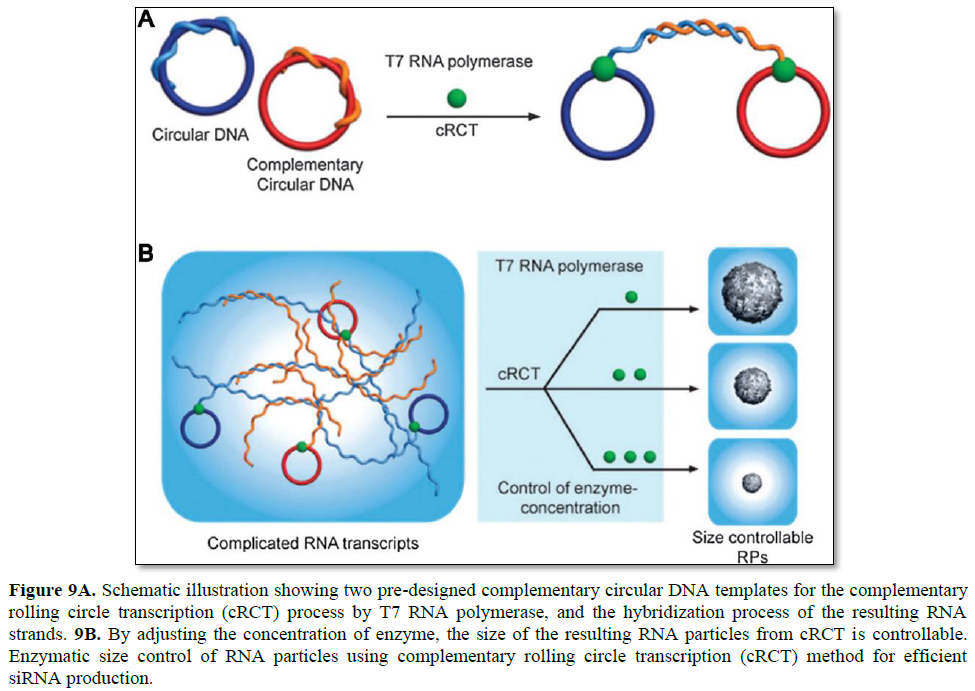
Since RCT has drawn a great deal of attention, the complementary rolling circle transcription (cRCT) method also emerged. Without any assistance from a synthetic polycation for condensing, enzymatic size control was feasible with a recently introduced cRCT method [28]. Extension of RNA strands from two complementary circular DNAs results in two strands that hybridize with each other, leading to the formation of RNA particles (Figure 9). Resulting double-stranded RNA is rationally designed to function as a substrate for the dicer enzyme to induce RNA interference. In the synthesis of RNA particles, T7 RNA polymerase is used to generate RNA strands, and the concentration of the enzymewas found to be critical to the size of the resulting RNA particles. By controlling the concentration of the enzyme in the cRCT reaction, the size of RNA nanoparticles was shrunk from 5 μm to 600 nm in diameter.
By taking advantage of cRCT, RNA membranes were also developed by Lee [29]. RNA membranes are designed to contain siRNA sequences to be cleaved by the dicer enzyme for siRNA release, and are the first example of a macroscopic RNA membrane composed solely of RNA strands. Furthermore, its structural and functional properties can be rationally controlled by adjusting the RNA base pairing, thus controlled release of chemical molecules and sequence-specific drug release are feasible.


Microscopic DNA scaffolds for gene delivery
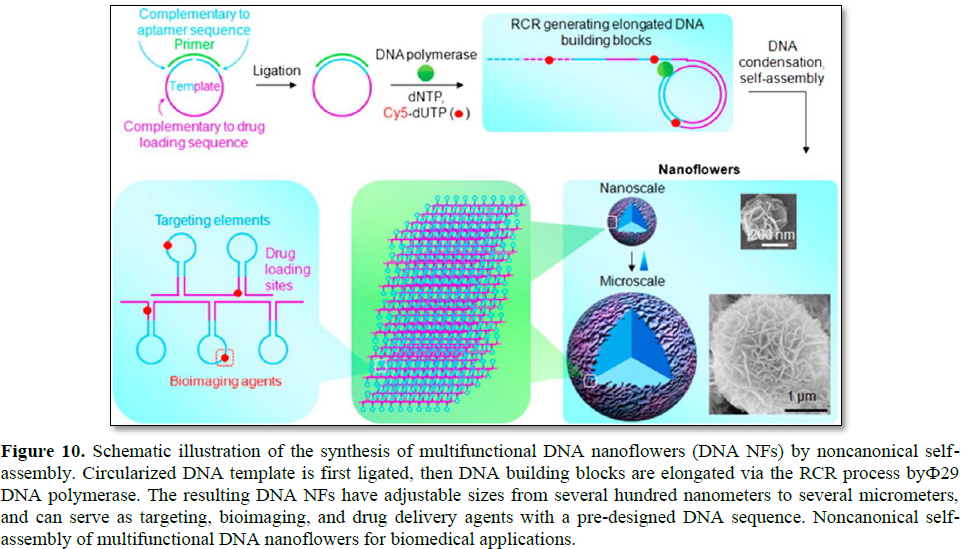
By utilizing RCR, DNA structures for gene delivery applications have also been introduced. For instance, self-assembled hierarchical DNA nanoflowers (NFs) with densely packed DNA and built-in multifunctional moieties
for versatile biomedical applications were developed by W. Tan's research group (Figure 10) [30]. The assembly of DNA NFs is independent of the traditional Watson-Crick base-pairing between DNA strands. Instead, it is driven by dense packaging of the resulting long building blocks generated via RCA and liquid crystallization, an anisotropic process for orderly alignment of highly concentrated polymers. In virtue of this distinctive characteristic, templates for RCA can be flexibly designed to include aptamers, antisense nucleotides, and drug loading sequences. Furthermore, the NFs were featured by sizetunability and stability towards nuclease treatment, dilution to low concentration, or denaturation by heating or urea treatment.
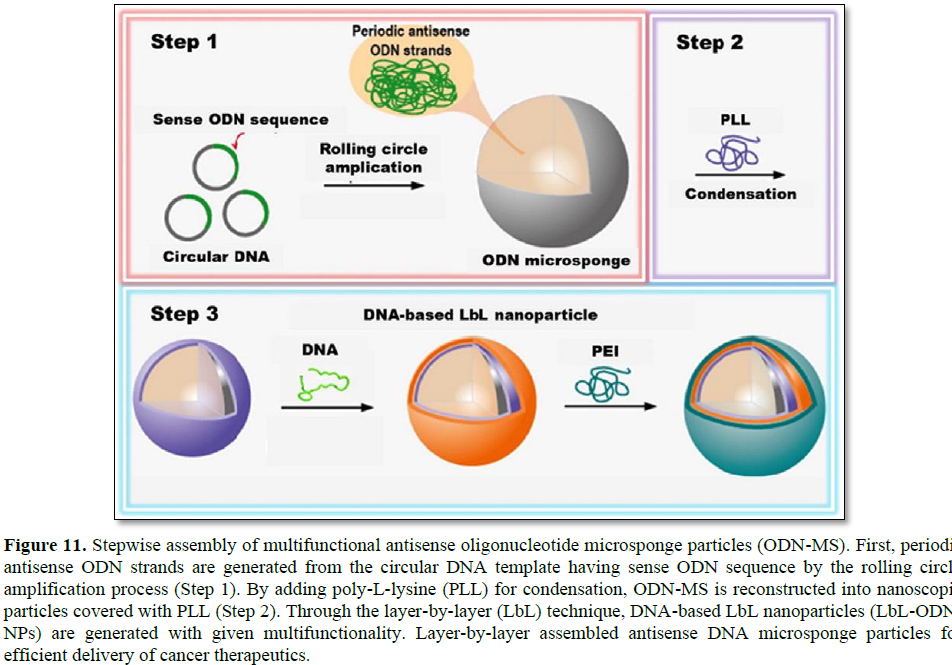
In addition, a layer-by-layer (LbL) assembly strategy offering a potent delivery method for nucleic acid therapeutics for cancer treatment was introduced by Hammond [31]. Oligonucleotide antisense microsponge particles (ODN-MS) were generated by RCA, then cationic polymers (poly-L-lysine: PLL) were added for condensing (Figure 11). By electrostatic interaction and physical agitation, the average size of ODN-MS decreased from 2 μm to 200 nm, which is a desirable size for efficient cellular uptake. Condensing with PLL also increased the stability of the ODN particles and changed the surface charge from negative to positive. Similarly, additional layers such as ssDNA and PEI could also be achieved by electrostatic interaction. The LbL assembly has advantages for controlling surface charge and fine-tuning of their multifunctional properties. This study implies that the spherical ODN nanoparticles could be nucleic acid carriers for gene delivery applications with negligible cytotoxicity. Moreover, this approach can achieve multi-functionality by adding layers of different functional DNAs and selecting different biomaterials.
Another novel approach to synthesize DNA structures is a cocoonlike self-degradable anticancer drug system that fully utilizes the RCA mechanism [118]. RCA was carried out with the template DNA that incorporates a palindromic sequence to facilitate self-assembly. The elongated DNA strands have multiple GC-pair sequences, achieving a high doxorubicin (DOX) loading capacity. Furthermore, this




bioinspired drug delivery carrier is functionalized by folic acid, and pH-responsive polymeric nano capsules encapsulate DNase I. Folic acid conjugated to the surface of the carrier promotes internalization of nanoclew into target cells and enters the acidic end lysosome. Subsequently, the acidic cellular environment activates the degradation of nano capsules embedded into the nano clew, thus triggering escape of DNase I from the nanocage. As a result, the DNA-based delivery vehicle can release intercalated DOX in target cells due to degradation of DNA by DNase I. This study demonstrated that RCA could be applied to long DNA synthesis, facilitating self-assembly of DNA-based structures. As a further step, the DNA structure was improved by adopting simple methods such as controlling DNA sequences and combining DNA with targetable or stimuli-responsive materials.
DNA nanoribbon (DNR), having a periodically repeating sequence, was also synthesized via RCA by Y. Weizmann's group [119]. While the conventional DNA origami strategy involves hundreds of staple strands, only three short staple strands are required in RCA-based DNR with three staple stands (DNR-T) production. Furthermore, because staple strands and scaffold strands are sequentially assembled by incorporating nicking processes into RCA, it is possible to generate DNR in a one-pot process. Despite the negative electric charge of DNR, its unique rigidity and ribbon-like structure enable DNR to easily pass through the cell membrane. Moreover, the rigidity of DNR is maintained after penetration, and the high aspect ratio allows DNR to easily escape from endosomal entrapment. In virtue of these characteristics, DNR can be used for effective siRNA delivery. siRNA-DNR-T, which has a high loading capacity, can deliver siRNA into the cytoplasm without requiring the proton sponge mechanism, and effectively mediates gene silencing in human cancer cells.
DNA/RNA NANOPARTICLES
DNA or RNA nanotechnology is the design, construction, and application of nucleic acid nanostructures using specific base paring and programmability of nucleic acids [120,121]. The bottom up self-assembly based on DNA/RNA nanotechnology has been used for various therapeutic applications [82].
pRNA Nanoparticles
Guo group has constructed RNA nanoparticles based on packaging RNA (pRNA) engineering and tried to apply the RNA nanoparticles to biomedical applications through functionalization with therapeutic molecules. pRNA is a component of bacteriophage phi29 DNA packing motor possessing two distinct domains. One is a dsRNA helical domain with 3′- and 5′-ends and the other is interlocking domain with two loops (right hand and left hand) [122]. The two loops are complementary to each other, which enables the pRNA to form dimer, trimer, and oligomeric structures by intermolecular interactions.
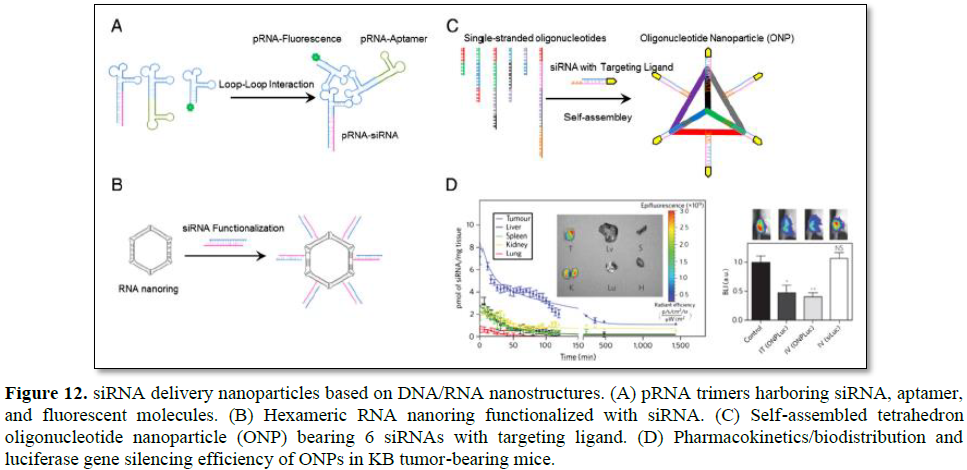
Through utilization of interlocking loops, trimeric pRNA nanoparticles were prepared (Figure 12A) [123]. Chimeric pRNAs were produced by replacing the helical domain with siRNA, aptamer, or folate. The reengineering of the helical domain did not hinder formation of trimeric pRNA nanoparticles. Trimeric pRNA nanoparticles harboring pRNAsiRNA, pRNA-CD4 aptamer, and pRNA-fluorescent molecule were treated to CD4 overexpressing cells to examine target specific codelivery of all the components at the same time. All the three functional molecules were target specifically delivered at one time and showed gene silencing activities. Dicer treatment of chimeric pRNA-siRNA also resulted in ~21 nt siRNA, which suggested that gene silencing was achieved through intracellular Dicer processed siRNAs. The advantage of trimeric pRNA nanoparticle is the ability to carry multiple siRNAs or other functional moieties at the same time.
RNA nanoring
RNA nanoring’s functionalized with siRNAs were constructed with high yield using RNAI and RNAII modules [22,124]. RNAI and RNAII are transcripts of plasmid that control the replication of ColE1 plasmid of Escherichia coli [125]. The RNAI/II can form so called an inverse kissing-loop complex, mediated by specific loop-loop interaction. Shapiro group carefully designed 6 loop sequence-modified RNA modules that were able to self-assemble into ~15 nm hexameric nanoring (Figure 12B). The RNA nanoring further functionalized with 6 siRNAs to use as a siRNA delivery system. Dicer treatment of the nanoring harboring siRNA produced siRNA of ~21 nt in length which could enter RNAi pathway. When the RNA nanoring with siRNAs was transfected into cells, efficient target gene inhibition was achieved. Because 6 siRNAs could be incorporated into a single RNA nanoring, multiple genes could be targeted at the same time for synergistic effects or 6 different regions of one gene could be targeted for more efficient gene silencing. Precise stoichiometric control of siRNA is another attractive quality of RNA nanotechnology-based delivery system.
Tetrahedron oligonucleotide nanoparticles
Conventional drug delivery systems based on polymers or lipids have heterogeneous size, shape, and composition which make it difficult to predict in vivo pharmacokinetics and pharmacodynamics of drugs [126]. To overcome these limitations, Lee et al. proposed siRNA delivery nanoparticles which are molecularly identical in size and shape by utilizing DNA nanotechnology [27]. Carefully designed 6 DNA strands were self-assembled into homogeneous tetrahedron oligonucleotide nanoparticle (ONP) using sequence-specific complementary base paring. Each edge of the ONP had a single stranded overhang which was complementary to overhang sequence of siRNA thereby 6 siRNAs can be accommodated in a single ONP (Figure 12C). The ONP showed a homogeneous structure of ~26.8 nm in size when measured using dynamic light scatter (DLS). This size of ONP was expected not only to be accumulated near tumor tissues by taking advantage of EPR effects but also to avoid renal clearance. ONPs harboring 6 siRNAs which were functionalized with folate at the end were treated into folate overexpressing KB cells in vitro without any cationic carriers. They exhibited efficient gene silencing in spite of the absence of cationic carriers. ONPs were intravenously injected into KB xenograft mouse to examine in vivo behavior of the particles. ONPs showed ~4 times longer blood circulation time than parental siRNA and mainly accumulated in the tumor site (Figure 12D). Furthermore, effective target gene silencing in the tumor was achieved without cationic carriers and no significant increased production of INF-α was observed. The influence of targeting ligand spatial orientation and density on cellular uptake and gene silencing was also investigated by taking advantage of precise structural control of ONPs. Although it is important to understand the relationship between cellular uptake and ligand density and orientation, it was difficult to figure out experimentally with conventional nanoparticles.
The DNA/RNA nanotechnology-based siRNA delivery particles have several outstanding features than conventional polymeric or liposomal delivery systems. First, uniform size and shape of nanostructures could be constructed on demand. Second, stoichiometry and geometry of ligands or therapeutic molecules can be precisely controlled. Third, multiple siRNAs could be incorporated in one nanoparticle for targeting multiple genes or several regions of one gene, which will improve therapeutic efficacy. Lastly, DNA and RNA are biomaterials and considered less toxic and immunogenic. Although there are some challenges, such as endosomal escape, production cost, stability, and more precise understanding of DNA/RNA folding, the DNA/RNA nanostructure-based particles are promising new type of delivery systems.


CONCLUSIONS
RNAi therapeutics has unique advantages over conventional pharmaceutical drugs. RNA interference is an endogenous gene regulation process, thus almost all genes can be modulated by siRNAs. The identification and selection of highly potent siRNA sequences has already been accomplished for many gene targets, and the synthesis of siRNAs on a large scale has been achieved. In addition, RNAi therapeutics has demonstrated promise in the treatment of cancers, viral diseases, and genetic disorders. Although significant progress has been made in the field of siRNA delivery, there remain challenges to be overcome. These challenges include 1) the minimization of off-target effects and immune stimulation, 2) target-specific accumulation of RNAi therapeutics after systemic administration, and 3) the induction of a potent RNAi effect at an acceptable dose level. The key to therapeutic achievement using an RNAi approach depends on delivery issues, thus advanced delivery strategies are critical to fully optimize the power of siRNAs.
Engineered design of synthetic DNA/RNA molecules [127-130] can generate pre-defined structures that can easily self-assemble to form nanoparticles with multiple functionalities. The field of oligonucleotide-based nanotechnology for biomedical applications is just emerging, but will play an important role in the delivery of siRNA. In particular, oligonucleotide-based structural RNAi systems described in this chapter are promising as a new generation of gene delivery carriers for cancer therapy. To realize clinical application of structural RNAi systems, the potency of the delivery systems needs to be optimized. One of the solutions may be the incorporation of highly specific ligands within the system. Preclinical data from various biopharmaceutical companies have suggested that the delivery of ligand-conjugated siRNA can be highly improved by the utilization of the engineered design of structural RNAi systems [131]. Another considerable issue in the delivery of structural RNAi systems is the facilitated endosomal release of these materials. It is important to understand the endosomal escape mechanism of structural RNAi systems and endeavor to use the endolytic properties to accelerate the transfer of active siRNAs into the cytoplasm. Future prospects of multimerized/branched siRNA structures and oligonucleotide-based structural RNAi systems with defined size and functionality will continue to improve the precision and efficacy of siRNA delivery.
-
Hammond SM, Bernstein E, Beach D, Hannon GJ (2000) An RNA-directed nuclease mediates post-transcriptional gene silencing in Drosophila cells. Nature 404: 293-296.
-
Oh YK, Park TG (2009) siRNA delivery systems for cancer treatment. Adv Drug Deliv Rev 61: 850-862.
-
Holen T (2005) Mechanisms of RNAi: mRNA cleavage fragments may indicate stalled RISC. J RNAi Gene Silencing 1: 21-25.
-
Robb GB, Rana TM (2007) RNA helicase A interacts with RISC in human cells and functions in RISC loading. Mol Cell 26: 523-537.
-
Wang J, Lu Z, Wientjes MG, Au JLS (2010) Delivery of siRNA therapeutics: Barriers and carriers. AAPS J 12: 492-503.
-
Lee RJ, Low PS (1994) Delivery of liposomes into cultured KB cells via folate receptor mediated endocytosis. J Biol Chem 269: 3198-3204.
-
Yang Y, Chen H, Vlahov IR, Cheng JX, Low PS (2007) Characterization of the pH of folate receptor-containing endosomes and the rate of hydrolysis of internalized acid labile folate-drug conjugates. J Pharmacol Exp Ther 321: 462-468.
-
Patil SD, Rhodes DG, Burgess DJ (2005) DNA-based therapeutics and DNA delivery systems: A comprehensive review. AAPS J 7: E61-E77.
-
Liu YP, Berkhout B (2011) miRNA cassettes in viral vectors: Problems and solutions. Biochim Biophys Acta 1809: 732-745.
-
Burnett JC, Rossi JJ RNA-based therapeutics: Current progress and future prospects. Chem Biol 19: 60-71.
-
Lee SH, Chung BH, Park TG, Nam YS, Mok H (2012) Small-interfering RNA (siRNA)-based functional micro- and nanostructures for efficient and selective gene silencing. Acc Chem Res 45: 1014-1025.
-
Andey T, Marepally S, Patel A, Jackson T, Sarkar S, et al. (2014) Cationic lipid guided short-hairpin RNA interference of annexin A2 attenuates tumor growth and metastasis in a mouse lung cancer stem cell model. J Control Release 184: 67-78.
-
Lee SY, Huh MS, Lee S, Lee SJ, Chung H, et al. (2010) Stability and cellular uptake of polymerized siRNA (poly-siRNA)/ polyethylenimine (PEI) complexes for efficient gene silencing. J Control Release 141: 339-346.
-
Mok H, Lee SH, Park JW, Park TG (2010) Multimeric small interfering ribonucleic acid for highly efficient sequence-specific gene silencing. Nat Mater 9: 272-278.
-
Lee SH, Mok H, Jo S, Hong CA, Park TG (2011) Dual gene targeted multimeric siRNA for combinatorial gene silencing. Biomaterials 32: 2359-2368.
-
Nakashima Y, Abe H, Abe N, Aikawa K, Ito Y (2011) Branched RNA nanostructures for RNA interference. Chem Commun 47: 8367-8369.
-
Hong CA, Lee SH, Kim JS, Park JW, Bae KH, et al. (2011) Gene silencing by siRNA microhydrogels via polymeric nanoscale condensation. J Am Chem Soc 133: 13914-13917.
-
Zhou J, Shu Y, Guo P, Smith DD, Rossi JJ (2011) Dual functional RNA nanoparticles containing phi29 motor pRNA and anti-gp120 aptamer for cell-type specific delivery and HIV-1 inhibition. Methods 54: 284-294.
-
Yoo H, Jung H, Kim SA, Mok H (2014) Multivalent comb-type aptamer-siRNA conjugates for efficient and selective intracellular delivery. Chem Commun 50: 6765-6767.
-
Grabow WW, Zakrevsky P, Afonin KA, Chworos A, Shapiro BA, et al. (2011) Self-assembling RNA nanoring’s based on RNAI/II inverse kissing complexes. Nano Lett 11: 878-887.
-
Afonin KA, Grabow WW, Walker FM, Bindewald E, Dobrovolskaia MA, et al. (2011) Design and self-assembly of siRNA-functionalized RNA nanoparticles for use in automated nanomedicine. Nat Protoc 6: 2022-2034.
-
Afonin KA, Kireeva M, Grabow WW, Kashlev M, Jaeger L, et al. (2012) Co-transcriptional assembly of chemically modified RNA nanoparticles functionalized with siRNAs. Nano Lett 12: 5192-5195.
-
Shu Y, Haque F, Shu D, Li W, Zhu Z, et al. (2013) Fabrication of 14 different RNA nanoparticles for specific tumor targeting without accumulation in normal organs. RNA 19: 767-777.
-
Haque F, Shu D, Shu Y, Shlyakhtenko LS, Rychahou PG, et al. (2012) Ultrastable synergistic tetravalent RNA nanoparticles for targeting to cancers. Nano Today 7: 245-257.
-
Shu D, Moll WD, Deng Z, Mao C, Guo P (2004) Bottom-up assembly of RNA arrays and superstructures as potential parts in nanotechnology. Nano Lett 4: 1717-1723.
-
Khaled A, Guo S, Li F, Guo P (2005) Controllable self-assembly of nanoparticles for specific delivery of multiple therapeutic molecules to cancer cells using RNA nanotechnology. Nano Lett 5: 1797-1808.
-
Lee H, Lytton-Jean AK, Chen Y, Love KT, Park AI, et al. (2012) Molecularly self-assembled nucleic acid nanoparticles for targeted in vivo siRNA delivery. Nat Nanotechnol 7: 389-393.
-
Han D, Park Y, Nam H, Lee JB (2014) Enzymatic size control of RNA particles using complementary rolling circle transcription (cRCT) method for efficient siRNA production. Chem Commun 50: 11665-11667.
-
Han D, Park Y, Kim H, Lee JB Self-assembly of free-standing RNA membranes. Nat Commun 5: 4367.
-
Zhu G, Hu R, Zhao Z, Chen Z, Zhang X, et al. (2013) Noncanonical self-assembly of multifunctional DNA nanoflowers for biomedical applications. J Am Chem Soc 135: 16438-16445.
-
Roh YH, Lee JB, Shopsowitz KE, Dreaden EC, Morton SW, et al. (2014) Layer-by-layer assembled antisense DNA microsponge particles for efficient delivery of cancer therapeutics. ACS Nano 8: 9767-9780.
-
Hannon GJ (2002) RNA interference. Nature 418(6894): 244-251.
-
Sharp PA (1999) RNAi and double-strand RNA. Gene Dev 13(2): 139-141.
-
Elbashir SM, Harborth J, Lendeckel W, Yalcin A, Weber K, et al. (2001) Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature 411(6836): 494-498.
-
Bernstein E, Caudy AA, Hammond SM, Hannon GJ (2001) Role for a bidentate ribonuclease in the initiation step of RNA interference. Nature 409(6818): 363-366.
-
Check E (2005) Acrucial test. Nat Med 11(3): 243-244.
-
DeVincenzo J, Cehelsky JE, Alvarez R, Elbashir S, Harborth J, et al. (2008) Evaluation of the safety, tolerability and pharmacokinetics of ALN-RSV01, a novel RNAi antiviral therapeutic directed against respiratory syncytial virus (RSV). Antiviral Res 77(3): 225-231.
-
Aagaard L, Rossi JJ (2007) RNAi therapeutics: Principles, prospects and challenges. Adv Drug Deliv Rev 59(2-3): 75-86.
-
Yuan X, Naguib S, Wu Z (2011) Recent advances of siRNA delivery by nanoparticles. Expert Opin Drug Deliv 8(4): 521-536.
-
Burnett JC, Rossi JJ, Tiemann K (2011) Current progress of siRNA/shRNA therapeutics in clinical trials. Biotechnol J 6(9): 1130-1146.
-
Liu YP, Berkhout B (2011) miRNA cassettes in viral vectors: Problems and solutions. Biochim Biophys Acta 1809(11-12): 732-745.
-
Sinn PL, Sauter SL, McCray Jr PB (2005) Gene therapy progress and prospects: Development of improved lentiviral and retroviral vectors-design, biosafety, and production. Gene Ther 12(14): 1089-1098.
-
Wang Y, Li Z, Han Y, Liang LH, Ji A (2010) Nanoparticle based delivery system for application of siRNA in vivo. Curr Drug Metab 11(2): 182-196.
-
Heidel JD, Yu Z, Liu JY, Rele SM, Liang Y, et al. (2007) Administration in non-human primates of escalating intravenous doses of targeted nanoparticles containing ribonucleotide reductase subunit M2 siRNA. Proc Natl Acad Sci U S A 104: 5715-5721.
-
Castanotto D, Rossi JJ (2009) The promises and pitfalls of RNA-interference-based therapeutics. Nature 457: 426-433.
-
Judge AD, Sood V, Shaw JR, Fang D, McClintock K, et al. (2005) Sequence dependent stimulation of the mammalian innate immune response by synthetic siRNA. Nat Biotechnol 23: 457-462.
-
Kim SH, Jeong JH, Lee SH, Kim SW, Park TG (2008) Local and systemic delivery of VEGF siRNA using polyelectrolyte complex micelles for effective treatment of cancer. J Control Release 129: 107-116.
-
Bolcato-Bellemin AL, Bonnet ME, Creusat G, Erbacher P, Behr JP (2007) Sticky overhangs enhance siRNA-mediated gene silencing. Proc Natl Acad Sci U S A 104: 16050-16055.
-
J. Gary, N. Puri, Y.Y. Won, Polymer-based siRNA delivery: Perspectives on the fundamental and phenomenological distinctions from polymer-based DNA delivery, J. Control. Release 121 (2007) 64–73.
-
Shah SA, Brunger AT (1999) The 1.8 A crystal structure of a statically disordered 17 base-pair RNA duplex: principles of RNA crystal packing and its effect on nucleic acid structure. J Mol Biol 285: 1577-1588.
-
Kebbekus P, Draper DE, Hagerman P (1995) Persistence length of RNA. Biochemistry 34: 4354-4357.
-
Tumova S, Woods A, Couchman JR (2000) Heparan sulfate proteoglycans on the cell surface: Versatile coordinators of cellular functions. Int J Biochem Cell Biol 32: 269-288.
-
Derfus AM, Chen AA, Min DH, Ruoslahti E, Bhatia SN (2007) Targeted quantum dot conjugates for siRNA delivery. Bioconjug Chem 18: 1391-1396.
-
Kim SH, Jeong JH, Lee SH, Kim SW, Park TG (2006) PEG conjugated VEGF siRNA for anti-angiogenic gene therapy. J Control Release 116: 123-129.
-
Jeong JH, Mok H, Oh YK, Park TG (2009) siRNA conjugate delivery systems. Bioconjug Chem 20: 5-14.
-
Chung HJ, Hong CA, Lee SH, Jo SD, Park TG (2011) Reducible siRNA dimeric conjugates for efficient cellular uptake and gene silencing. Bioconjug Chem 22: 299-306.
-
Lee SJ, Son S, Yhee JY, Choi K, Kwon IC, et al. (2013) Structural modification of siRNA for efficient gene silencing. Biotechnol Adv 31: 491-503.
-
Kong WH, Bae KH, Hong CA, Lee Y, Hahn SK, et al. (2011) Multimerized siRNA cross-linked by gold nanoparticles. Bioconjug Chem 22: 1962-1969.
-
Alivisatos AP, Johnsson KP, Peng X, Wilson TE, Loweth CJ, et al. (1996) Organization of 'nanocrystal molecules' using DNA. Nature 382: 609-611.
-
Mirkin CA, Letsinger RL, Mucic RC, Storhoff JJ (1996) A DNA-based method for rationally assembling nanoparticles into macroscopic materials. Nature 382: 607-609.
-
Aldaye FA, Sleiman HF (2007) Dynamic DNA templates for discrete gold nanoparticle assemblies: control of geometry, modularity, write/erase and structural switching. J Am Chem Soc 129: 4130-4131.
-
Ghosh P, Han G, De M, Kim CK, Rotello VM (2008) Gold nanoparticles in delivery applications. Adv Drug Deliv Rev 60: 1307-1315.
-
Gao K, Huang L (2009) Nonviral methods for siRNA delivery. Mol Pharm 6: 651-658.
-
Lv H, Zhang S, Wang B, Cui S, Yan J (2006) Toxicity of cationic lipids and cationic polymers in gene delivery. J Control Release 114: 100-109.
-
Lee SJ, Huh MS, Lee SY, Min S, Lee S, et al. (2012) Tumor-homing poly-siRNA/glycol chitosan self-cross-linked nanoparticles for systemic siRNA delivery in cancer treatment. Angew Chem 51: 7203-7207.
-
Yhee JY, Lee SJ, Lee S, Song S, Min HS, et al. (2013) Tumor-targeting transferrin nanoparticles for systemic polymerized siRNA delivery in tumor-bearing mice. Bioconjug Chem 24: 1850-1860.
-
Kuijpers AJ, van Wachem PB, van Luyn MJ, Brouwer LA, Engbers GH, et al. (2000) In vitro and in vivo evaluation of gelatin chondroitin sulphate hydrogels for controlled release of antibacterial proteins. Biomaterials 21: 1763-1772.
-
Ishikawa H, Nakamura Y, Jo J, Tabata Y (2012) Gelatin nanospheres incorporating siRNA for controlled intracellular release. Biomaterials 33: 9097-9104.
-
Son S, Song S, Lee SJ, Min S, Kim SA, et al. (2013) Self-crosslinked human serum albumin nanocarriers for systemic delivery of polymerized siRNA to tumors. Biomaterials 34: 9475-9485.
-
Whitehead KA, Langer R, Anderson DG (2009) Knocking down barriers: advances in siRNA delivery. Nat Rev Drug Discov 8: 129-138.
-
Boado RJ (2007) Blood-brain barrier transport of non-viral gene and RNAi therapeutics. Pharm Res 24: 1772-1787.
-
LaGrandeur TE, Huttenhofer A, Noller HF, Pace NR (1994) Phylogenetic comparative chemical footprint analysis of the interaction between ribonuclease P RNA and tRNA. EMBO J 13: 3945-3952.
-
Chen C, Zhang C, Guo P (1999) Sequence requirement for hand-in-hand interaction in formation of RNA dimers and hexamers to gear phi29 DNA translocation motor. RNA 5: 805-818.
-
Afonin KA, Kasprzak WK, Bindewald E, Kireeva M, Viard M, et al. (2014) Shapiro, In silico design and enzymatic synthesis of functional RNA nanoparticles. Acc Chem Res 47: 1731-1741.
-
Afonin KA, Bindewald E, Yaghoubian AJ, Voss N, Jacovetty E, et al. (2010) In vitro assembly of cubic RNA-based scaffolds designed in silico. Nat Nanotechnol 5: 676-682.
-
Severcan I, Geary C, Chworos A, Voss N, Jacovetty E, et al. (2010) A polyhedron made of tRNAs. Nat Chem 2: 772-779.
-
Grabow WW, Jaeger L (2014) RNA self-assembly and RNA nanotechnology. Acc Chem Res 47: 1871-1880.
-
Chworos A, Severcan I, Koyfman AY, Weinkam P, Oroudjev E, et al. (2004) Building programmable jigsaw puzzles with RNA. Science 306: 2068-2072.
-
Guo P (2010) The emerging field of RNA nanotechnology. Nat Nanotechnol 5: 833-842.
-
Shukla GC, Haque F, Tor Y, Wilhelmsson LM, Toulme JJ, et al. (2011) A boost for the emerging field of RNA nanotechnology. ACS Nano 5: 3405-3418.
-
Koyfman AY, Braun G, Magonov S, Chworos A, Reich NO, et al. (2005) Controlled spacing of cationic gold nanoparticles by nanocrown RNA. J Am Chem Soc 127: 11886-11887.
-
Shu Y, Pi F, Sharma A, Rajabi M, Haque F, et al. (2014) Stable RNA nanoparticles as potential new generation drugs for cancer therapy. Adv Drug Deliv Rev 66: 74-89.
-
Khisamutdinov EF, Jasinski DL, Guo P (2014) RNA as a boiling-resistant anionic polymer material to build robust structures with defined shape and stoichiometry. ACS Nano 8: 4771-4781.
-
Hao C, Li X, Tian C, Jiang W, Wang G, et al. (2014) Construction of RNA nanocages by re-engineering the packaging RNA of Phi29 bacteriophage. Nat Commun 5: 3890.
-
Ohno H, Kobayashi T, Kabata R, Endo K, Iwasa T, et al. (2011) Synthetic RNA-protein complex shaped like an equilateral triangle. Nat Nanotechnol 6: 116-120.
-
Osada E, Suzuki Y, Hidaka K, Ohno H, Sugiyama H, et al. (2014) Engineering RNA-protein complexes with different shapes for imaging and therapeutic applications. ACS Nano 8: 8130-8140.
-
Tarapore P, Shu Y, Guo P, Ho SM (2011) Application of phi29 motor pRNA for targeted therapeutic delivery of siRNA silencing metallothionein-IIA and survivin in ovarian cancers. Mol Ther 19: 386-394.
-
Petros RA, DeSimone JM (2010) Strategies in the design of nanoparticles for therapeutic applications. Nat Rev Drug Discov 9: 615-627.
-
Kaur J, Tikoo K (2015) Ets1 identified as a novel molecular target of RNA aptamer selected against metastatic cells for targeted delivery of nano-formulation. Oncogene 34: 5216-5228.
-
Baneyx G, Baugh L, Vogel V (2002) Fibronectin extension and unfolding within cell matrix fibrils controlled by cytoskeletal tension. Proc Natl Acad Sci U S A 99: 5139-5143.
-
Hyman P, Valluzzi R, Goldberg E (2002) Design of protein struts for self-assembling nanoconstructs. Proc Natl Acad Sci U S A 99: 8488-8493.
-
Goldberger J, He R, Zhang Y, Lee S, Yan H, et al. (2003) Single-crystal gallium nitride nanotubes. Nature 422: 599-602.
-
Shu D, Shu Y, Haque F, Abdelmawla S, Guo P (2011) Thermodynamically stable RNA three-way junction for constructing multifunctional nanoparticles for delivery of therapeutics. Nat Nanotechnol 6: 658-667.
-
Shu Y, Cinier M, Fox SR, Ben-Johnathan N, Guo P (2011) Assembly of therapeutic pRNA siRNA nanoparticles using bipartite approach. Mol Ther 19: 1304-1311.
-
Goodman RP, Schaap IA, Tardin CF, Erben CM, Berry RM, et al. (2005) Rapid chiral assembly of rigid DNA building blocks for molecular nanofabrication. Science 310: 1661-1665.
-
He Y, Ye T, Su M, Zhang C, Ribbe AE, et al. (2008) Hierarchical self-assembly of DNA into symmetric supramolecular polyhedra. Nature 452: 198-201.
-
Bhatia D, Surana S, Chakraborty S, Koushika SP, Krishnan Y (2011) A synthetic icosahedral DNA-based host-cargo complex for functional in vivo imaging. Nat Commun 2: 339.
-
Walsh AS, Yin H, Erben CM, Wood MJ, Turberfield AJ (2011) DNA cage delivery to mammalian cells. ACS Nano 5: 5427-5432.
-
Keum JW, Ahn JH, Bermudez H (2011) Design, assembly, and activity of antisense DNA nanostructures. Small 7: 3529-3535.
-
Gaglione M, Messere A (2010) Recent progress in chemically modified siRNAs. Mini Rev Med Chem 10: 578-595.
-
Li Z, Wei B, Nangreave J, Lin C, Liu Y, et al. (2009) A replicable tetrahedral nanostructure self-assembled from a single DNA strand. J Am Chem Soc 131: 13093-13098.
-
Fire A, Xu SQ (1995) Rolling replication of short DNA circles. Proc Natl Acad Sci U S A 92: 4641-4645.
-
Liu D, Daubendiek SL, Zillman MA, Ryan K, Kool ET (1996) Rolling circle DNA synthesis: Small circular oligonucleotides as efficient templates for DNA polymerases. J Am Chem Soc 118: 1587-1594.
-
Lin C, Xie M, Chen JJ, Liu Y, Yan H (2006) Rolling-circle amplification of a DNA nanojunction. Angew Chem 45: 7537-7539.
-
Hong CA, Jang B, Jeong EH, Jeong H, Lee H (2014) Self-assembled DNA nanostructures prepared by rolling circle amplification for the delivery of siRNA conjugates. Chem Commun 50: 13049-13051.
-
Ali MM, Li F, Zhang Z, Zhang K, Kang DK, et al. (2014) Rolling circle amplification: A versatile tool for chemical biology, materials science and medicine. Chem Soc Rev 43: 3324-3341.
-
Daubendiek SL, Ryan K, Kool ET (1995) Rolling-circle RNA synthesis: Circular oligonucleotides as efficient substrates for T7 RNA polymerase. J Am Chem Soc 117: 7818-7819.
-
Zhang D, Wu J, Ye F, Feng T, Lee I, et al. (2006) Amplification of circularizable probes for the detection of target nucleic acids and proteins. Clinica chimica acta. Int J Clin Chem 363: 61-70.
-
Fischer NO, Tarasow TM, Tok JB (2008) Protein detection via direct enzymatic amplification of short DNA aptamers. Anal Biochem 373: 121-128.
-
Jarvius J, Melin J, Göransson J, Stenberg J, Fredriksson S, et al. (2006) Digital quantification using amplified single-molecule detection. Nat Methods 3: 725-727.
-
Larsson C, Grundberg I, Soderberg O, Nilsson M (2010) In situ detection and genotyping of individual mRNA molecules. Nat Methods 7: 395-397.
-
Zhang Z, Ali MM, Eckert MA, Kang DK, Chen YY, et al. (2013) A polyvalent aptamer system for targeted drug delivery. Biomaterials 34: 9728-9735.
-
Lee JB, Hong J, Bonner DK, Poon Z, Hammond PT (2012) Self-assembled RNA interference microsponges for efficient siRNA delivery. Nat Mater 11: 316-322.
-
Qi H, Ghodousi M, Du Y, Grun C, Bae H, et al. (2013) DNA-directed self-assembly of shape-controlled hydrogels. Nat Commun 4: 2275.
-
Lee JB, Peng S, Yang D, Roh YH, Funabashi H, et al. (2012) A mechanical metamaterial made from a DNA hydrogel. Nat Nanotechnol 7: 816-820.
-
Daubendiek SL, Kool ET (1997) Generation of catalytic RNAs by rolling transcription of synthetic DNA nanocircles. Nat Biotechnol 15: 273-277.
-
Seyhan AA, Vlassov AV, Johnston BH (2006) RNA interference from multimeric shRNAs generated by rolling circle transcription. Oligonucleotides 16: 353-363.
-
Sun W, Jiang T, Lu Y, Reiff M, Mo R, et al. (2014) Cocoon-like self-degradable DNA nanoclew for anticancer drug delivery. J Am Chem Soc 136: 14722-14725.
-
Chen G, Liu D, He C, Gannett TR, Lin W, et al. (2015) Enzymatic synthesis of periodic DNA nanoribbons for intracellular pH sensing and gene silencing. J Am Chem Soc 137: 3844-3851.
-
Pinheiro AV, Han DR, Shih WM, Yan H (2011) Challenges and opportunities for structural DNA nanotechnology. Nat Nanotechnol 6: 763-772.
-
Guo PX (2010) The emerging field of RNA nanotechnology. Nat Nanotechnol 5: 833-842.
-
Guo PX, Erickson S, Anderson D (1987) A small viral-RNA is required for in vitro packaging of bacteriophage-Phi-29 DNA. Science 236: 690-694.
-
Khaled A, Guo SC, Li F, Guo PX (2005) Controllable self-assembly of nanoparticles for specific delivery of multiple therapeutic molecules to cancer cells using RNA nanotechnology. Nano Lett 5: 1797-1808.
-
Grabow WW, Zakrevsky P, Afonin KA, Chworos A, Shapiro BA, et al. (2011) Self-assembling RNA nanoring’s based on RNAI/II inverse kissing complexes. Nano Lett 11: 878-887.
-
Eguchi Y, Tomizawa J (1990) Complex formed by complementary RNA stem-loops and its stabilization by a protein - function of Cole1 Rom protein. Cell 60: 199-209.
-
Petros RA, DeSimone JM (2010) Strategies in the design of nanoparticles for therapeutic applications. Nat Rev Drug Discov 9: 615-627.
-
Madkour LH (2020) Book: Reactive Oxygen Species (ROS), Nanoparticles, and Endoplasmic Reticulum (ER) Stress-Induced Cell Death Mechanisms. Paperback ISBN: 9780128224816. Published Date: August 1, 2020. Available online at: https://www.elsevier.com/books/reactive-oxygen-species-ros-nanoparticles-and-endoplasmic-reticulum-er-stress-induced-cell-death-mechanisms/madkour/978-0-12-822481-6
-
Madkour LH (2020) Book: Nanoparticles Induce Oxidative and Endoplasmic Reticulum Antioxidant Therapeutic Defenses. Copyright 2020 Publisher Springer International Publishing. Copyright Holder Springer Nature Switzerland AG eBook ISBN 978-3-030-37297-2, Hardcover ISBN 978-3-030-37296-5 Series ISSN 2194-0452 Edition Number 1. Available online at: https://www.springer.com/gp/book/9783030372965?utm_campaign=3_pier05_buy_print&utm_content=en_08082017&utm_medium=referral&utm_source=google_books#otherversion=9783030372972
-
Madkour LH (2020) Book: Nucleic Acids as Gene Anticancer Drug Delivery Therapy. 1st ed. Publishing house: Elsevier. Paperback ISBN: 9780128197776 Imprint: Academic Press Published Date: 2nd January 2020 Imprint: Academic Press Copyright: Paperback ISBN: 9780128197776 © Academic Press 2020 Published: 2nd January 2020 Imprint: Academic Press Paperback ISBN: Available online at: https://www.elsevier.com/books/nucleic-acids-as-gene-anticancer-drug-delivery%20therapy/madkour/978-0-12-819777-6
-
130. Madkour LH (2019) Book: Nanoelectronic Materials: Fundamentals and Applications (Advanced Structured Materials) 1st ed. Available online at: https://link.springer.com/book/10.1007%2F978-3-030-21621-4 Series Title Advanced Structured Materials Series Volume 116 Copyright 2019 Publisher Springer International Publishing Copyright Holder Springer Nature Switzerland AG eBook ISBN 978-3-030-21621-4, Hardcover ISBN 978-3-030-21620-7 Series ISSN 1869-8433 Edition Number 1 Number of Pages XLIII, 783 Number of Illustrations 122 b/w illustrations, 494 illustrations in color Topics ISBN-10: 3030216209 ISBN-13: 978-3030216207 #117 in Nanotechnology (Books) #725 in Materials Science (Books) #187336 in Textbooks https://books.google.com.eg/books/about/Nanoelectronic_Materials.html?id=YQXCxAEACAAJ&source=kp_book_description&redir_esc=y, https://www.springer.com/gp/book/9783030216207
-
Nair JK, Willoughby JL, Chan A, Charisse K, Alam MR, et al. (2014) Multivalent N-acetylgalactosamine-conjugated siRNA localizes in hepatocytes and elicits robust RNAi-mediated gene silencing. J Am Chem Soc 136: 16958-16961.