1179
Views & Citations179
Likes & Shares
DNA constitutes a
powerful target for chemotherapeutic intervention in human cancers,
particularly for those where high proliferation rates of tumor cell types have
resulted in sensitivity to drugs, which block replication and transcription of
their DNA. Molecular detection of particular DNA sites by small molecules is an
essential dilemma in drug design. Polycyclic heterocycles having a planar
structure can be efficient pharmacophore moieties for DNA-interactive drugs
because they can insert between the stacked bases paired oligonucleotides or
interact with grooves. Despite DNA being a significant target for numerous drugs,
most of the docking programs are validated only for proteins and their ligands.
In this paper, AutoDock 4.0 was used to perform self-dockings and cross
dockings between seven DNA targets and five ligands belonging to
1,2-Phenylenediamine Schiff’s base derivatives. AutoDock is able to correctly
recognize main DNA binding modes. The obtained docking results are in absolute
agreement with experimental data from the literature. In conclusion, our data
that computational approach on synthesized proposed ligands will contribute to
select the most promising candidates as DNA-interactive drugs that have
antitumor activity.
INTRODUCTION
DNA represents a major target for chemotherapeutic strategy in human
cancers, particularly for those where elevated proliferation rates of some
tumor cell types have resulted in sensitivity to drugs, which obstruct transcription
and replication of their DNA [1]. Molecular identification of DNA by small
molecules is an essential dilemma in drug design. Many polycyclic heterocyclic
having a planar structure could be efficient pharmacophore moieties for
DNA-interactive drugs since they are capable to insert themselves between the
stacked base paired oligonucleotides. Furthermore, if they have appropriate
side chains, additional interactions of these ligands with other important
architectural characteristic of DNA can be predicted [2,3]. We have recently
reported on using microwave assisted synthesis and antimicrobial evaluation of
symmetrical 1,2-Phenylenediamine Schiff’s base derivatives finding that some of
the synthesized compounds showed antibacterial and antifungal activity [4]. Now
we intended to explore how different structural features of
1,2-Phenylenediamine Schiff’s base derivatives can affect the DNA binding
capability. Here we present molecular modeling studies of 1,2-Phenylenediamine
Schiff’s base derivatives using seven DNA targets. We hypothesized that this
interaction can help to recognize the molecular mechanism of
1,2-Phenylenediamine Schiff’s base derivatives action and may serve as a basis
for understanding the molecular mechanism of action of the 1,2-Phenylenediamine
Schiff’s base derivatives and can help to design of new chemotherapeutic
molecules.
MATERIALS AND METHODS
Molecular docking
study
MGL (Molecular Graphics Laboratory) tools 1.5.4 with AutoDock4 and
AutoGrid 4.0 were used to set up and exert blind docking calculations between
various 1,2-Phenylenediamine Schiff’s base derivatives and DNA sequences. DNA
sequences:
DNA (5'-D (*CP*GP*CP*GP*AP*AP*TP*TP*CP*GP*CP*G)-3') (PDB ID: 1bna),
DNA (5'-D (*CP*GP*CP*AP*AP*AP*TP*TP*TP*GP*CP*G)-3') (PDB ID: 102d),
DNA (5'-D (*CP*GP*TP*AP*CP*G)-3') (PDB ID: 1k2j),
DNA (5'-D (*CP*GP*CP*GP*AP*TP*AP*TP*CP*GP*CP*G)-3') (PDB ID: 1dne),
DNA (5'-D (*CP*GP*CP*AP*GP*AP*AP*TP*TP*CP*GP*CP*G)-3') (PDB ID: 1d31),
DNA (5'-D (*CP*GP*CP*GP*AP*AP*TP*TP*CP*GP*CP*G)-3') (PDB ID: 2gvr), and
DNA (5'-D (*CP*GP*TP*AP*CP*G)-3') (PDB ID: 2des) were obtained from the
Protein Data Bank and were used for the docking studies. 1,2-Phenylenediamine
Schiff’s base derivatives structures were drawn and optimized using ChemDraw
Ultra (version 8.0, Cambridgesoft Com., USA). Chem3D Ultra was used to convert
2D into 3D structures and the energy was minimized using the semi-empirical AM1
method which is based on the Neglect of Differential Diatomic Overlap (NDDO)
integral approximation. The molecular dockings of SW (1,2-Phenylenediamine
Schiff’s Base derivatives) compounds with B-DNAs (B: right-handed double helix
DNA) were accomplished by Auto Dock 4.2 software from the Scripps Research
Institute (TSRI) (http://autodock.scripps.edu/). Firstly, the polar
hydrogen atoms were added into B-DNA molecules. Then, the partial atomic
charges of the B-DNA and SW molecules were calculated using Kollman methods
[5]. In the process of molecular docking, the grid maps of dimensions (62 Å ×
62 Å × 62 Å) with a grid-point spacing of 0.376Å and the grid boxes centered.
The number of genetic algorithm runs and the number of evaluations was set to
100. All other parameters were default settings. Cluster analysis was performed
on docking results by using a root mean square (RMS) tolerance of 2.0 Å,
dependent on the binding free energy. Lastly, the dominating configuration of
the binding complex of SW compounds and B-DNA fragments with minimum binding
energy can be determined. Taxol was used as a reference as it is a successful
drug that is used for the treatment of various cancers and it binds to DNA
grooves throughout the eight-membered taxane core ring, with the three phenyl
rings pointing away from the core eight-membered ring. In addition, taxol has
been reported to interact with tubulin leading to tumor cell death [6,7].
Binding energy
To molecular docking simulation method is primarily validated on basis of
the obtained binding energy. The predefined range of binding energy is supposed
to be in the range between -5 to -15 Kcal/mol to productively validate the
molecular docking process.
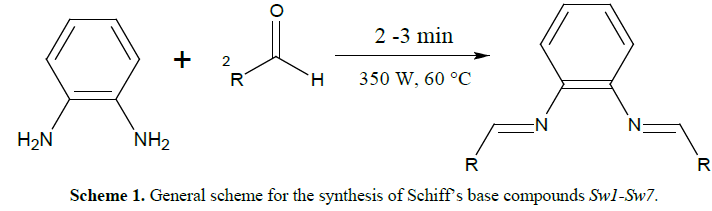
General procedure for
synthesis of Schiff’s bases (Sw1-Sw7)
[4]
The Schiff’s base was
prepared as described before by our group by reaction of one mole of
phenylenediamine and two moles of substituted aromatic aldehydes (Table 1)
[4]. All reactants were mixed together and a minimum amount of ethanol was
added (2-3 ml). This mixture was subjected to microwave irradiation at 350 W
for 2-3 min at 60°C. The development of the reaction was watched on thin layer
chromatography. The mixture was left for cooling and the solid product (crude)
was gathered by filtration and washed four times with ethanol and vacuum dried.
The gained product was re-dissolved in ethanol for recrystallization and dried
to give a pure product (Scheme 1). The crystalline products obtained
were characterized as published previously by FTIR, 1H NMR [4].
RESULTS AND DISCUSSION
Molecular docking
analysis
Table 2 shows the binding energies of SW compounds and DNAs fragments obtained
by the molecular docking strategy. In this study, molecular dockings of the SW
compounds with seven B-DNA fragments were performed using Auto Dock 4.2 to
investigate the binding mode of SW compounds with B-DNA and to obtain
information about interaction forces between SW compounds and DNA. SW compounds
and DNA were kept as flexible molecules and were docked into seven forms of
rigid B-DNA fragments to obtain the preferential binding site to SW compounds
on B-DNAs. The molecular docking results are shown in Table 2. The
modeling studies showed that there are van
der Waals, hydrogen bonding and electrostatic interactions between SW
compounds and DNAs. The contribution of van
der Waals and hydrogen bonding interaction is much greater than that of the
electrostatic interaction because the sum of van der Waals energy, hydrogen bonding energy and desolvation free
energy is larger than the electrostatic energy, which is consistent with the
literature [8,9].
Some of the binding energies obtained by performing molecular docking
simulation of the SW compounds with the seven DNA fragments did not lie in the
predefined range of -5 to -15 kcal/mol (Table 2). The obtained binding
energy results demonstrate that the affinity of SW compounds for their
‘‘preferred’’ sites is modulated by the local DNA sequence. In some cases this
effect is relatively small, while in others cases, as in SW5, SW6 and SW7 the
effect is dramatic. Since these sequence effects lie outside the principal
binding sites for these ligands, they may reflect changes in the local DNA
structure and/or dynamics. This is similar to those seen in protein-DNA
interactions [10,11].
Molecular docking is used
for virtual screening of SW compounds employing binding affinity and the best
orientation possible with respect to the target DNA. To illustrate the DNA-SW
compounds interactions an example from the series was chosen (SW7 compound).
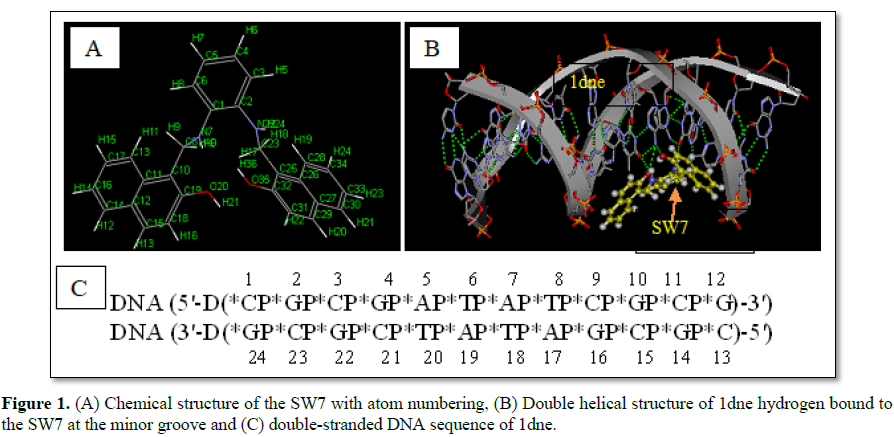
The SW7-DNA interactions are shown in Figure 1. SW7 compound showed a
good binding energy (-5.23 kcal/mol,) when compared to standard taxol (-0.69
kcal/mol) as mentioned in Table 2. The chemical structure of the SW7
with atom numbering is shown in Figure 1A. The double helical structure of 1dne
(Figure 1B), for example, is due largely to the hydrogen bonding between
the base pairs, which link one complementary strand to the other. Figure 1B
shows four hydrogen bonds between SW7 and 1dne fragment in the minor groove.
The four hydrogen bonds were AP5:N6 (of 1dne, as H-bond donor) and O20 (of SW7,
as H-bond acceptor); AP5:N6 (of 1dne, as H-bond donor) and O35 (of SW7, as
H-bond acceptor); H9 (of SW7, as H-bond donor) and TP20:O4 (of 1dne, as H-bond
acceptor); H24 (of SW7, as H-bond donor)and GP4:O6 (of 1dne, as H-bond
acceptor).
All SW compounds studied in this paper were bound to the minor groove of
the seven DNA fragments. This binding often has cytotoxic activity because they
interfere with the binding of proteins necessary for DNA replication and
transcription. In the literature compounds that bind to the minor groove of DNA
have proven to be very useful as antitumor agents because they selectively kill
rapidly-dividing cells [12-14]. This has encouraged efforts to design molecules
that bind at designated sites in the minor groove. It is thought that groove
binders with increased selectivity will produce a greater biological response
for a given dose (and consequently have fewer toxic and side effects) than
non-selective groove binders [12-14]. Molecules that target particular DNA
sites also have the prospective to be used for the selective suppression of
transcription from fastidious gene sequences [15]. The complexes of SW
compounds and DNA fragments could be stabilized by hydrogen bonding upon minor
groove binding. This assumption is confirmed by the literature studies as it
has been reported that the synthetic polycarboxamides consisting of
N-methyl-3-hydroxypyrrole (Hp), N-methylimidazole (Im), N-methylpyrrole (Py)
and beta-alanine (beta) showed strong and sequence-specific interaction with
the DNA minor groove when they form hairpin structures with side-by-side
antiparallel motifs [16,17]. In the synthetic polycarboxamides report the
researchers found new conjugates containing two ligands linked to the same
terminal phosphate of the DNA strand. The polycarboxamides are inserted into
the minor groove of a duplex in a parallel or antiparallel orientation. The
obtained stabilization of DNA duplexes by two attached minor groove ligands was
confirmed by thermal denaturation studies [16,17].
CONCLUSION
In conclusion, we have successfully performed in silico modelling for seven symmetrical 1,2-phenylenediamine
derivatives with seven DNA fragments. Relationships between hydrogen bond
geometry and positioning of the SW compounds with the minor groove were
studied. Docking studies suggest that molecular docking techniques may have
particular value as a virtual screening precursor step to full chemical
synthesis of drug candidates.
1.
Kumar R, Lown JW (2005) Design, synthesis and in vitro cytotoxic studies of novel
bis-pyrrolo[2,1][1,4] benzodiazepine-pyrrole and imidazole polyamide
conjugates. Eur J Med Chem 40: 641-654.
2.
Hu P, Chi HM, DeBacker KC, Gong X, Keim JH, et al.
(2019) Quaternary-centre-guided synthesis of complex polycyclic terpenes.
Nature.
3.
Thomas AM, He C, Zhao L, Galimova GR, Mebel AM, et
al. (2019) A combined experimental and computational study on the reaction
dynamics of the 1-propynyl (ch3cc) - 1,3-butadiene (CH2CHCHCH2)
System and the formation of toluene under single collision conditions. J Phys
Chem A.
4.
Mohamed SS, Al-Sadawi IA, Gbaj MA, Alsabri SG,
Elmaki NM, et al. (2018) Microwave assisted synthesis and antimicrobial
evaluation of symmetrical 1,2-phenylenediamine Schiff's base derivatives. Pharm
Pharmacol Int J 6: 344-348.
5.
Tiwari R, Mahasenan K, Pavlovicz R, Li C, Tjarks W
(2009) Carborane clusters in computational drug design: A comparative docking
evaluation using AutoDock, FlexX, Glide and Surflex. J Chem Inf Model 49:
1581-1589.
6.
Yu B, Tian X, Zhang L, Feng R (2016) Hematopoietic
PBX-interaction protein promotes breast cancer sensitivity to paclitaxel
through a microtubule-dependent mechanism. DNA Cell Biol 35: 740-745.
7.
Cheng HY, Zhang T, Qu Y, Shi WJ, Lou G, et al.
(2016) Synergism between RIZ1 gene therapy and paclitaxel in SiHa cervical
cancer cells. Cancer Gene Ther 23: 392-395.
8.
Holt PA, Chaires JB, Trent JO (2008) Molecular
docking of intercalators and groove-binders to nucleic acids using Autodock and
Surflex. J Chem Inf Model 48: 1602-1615.
9.
Gilad Y, Senderowitz H (2014) Docking studies on DNA
intercalators. J Chem Inf Model 54: 96-107.
10.
Hampshire AJ, Fox KR (2008) The effects of local DNA
sequence on the interaction of ligands with their preferred binding sites.
Biochimie 90: 988-998.
11.
Waring MJ (1986) Overview of the interaction between
chemotherapeutic agents and DNA. Drugs Exp Clin Res 12: 441-453.
12.
Hassan AA, Aly AA, Mohamed NK, El Shaieb KM,
Makhlouf MM, et al. (2019) Design, synthesis and DNA interaction studies of
furo-imidazo[3.3.3]propellane derivatives: Potential anticancer agents. Bioorg
Chem 85: 585-599.
13.
Mohan S, Rangappa S, Anilkumar NC, Fuchs JE, Bender
A, et al. (2019) Sulfated ceria catalyzed synthesis of imidazopyridines and
their implementation as DNA minor groove binders. Chem Biodivers.
14.
Carter EK, Laughlin-Toth S, Dodd T, Wilson WD,
Ivanov I (2019) Small molecule binders recognize DNA microstructural variations
via an induced fit mechanism. Phys Chem Chem Phys 21: 1841-1851.
15.
Ho SN, Boyer SH, Schreiber SL, Danishefsky SJ,
Crabtree GR (1994) Specific inhibition of formation of transcription complexes
by a calicheamicin oligosaccharide: A paradigm for the development of
transcriptional antagonists. Proc Natl Acad Sci U S A 91: 9203-9207.
16.
Ryabinin VA, Boutorine AS, Helene C, Denisov AY,
Pyshnyi DV, et al. (2004) Oligonucleotide--minor groove binder 1:2 conjugates:
Side by side parallel minor groove binder motif in stabilization of DNA duplex.
Nucleosides Nucleotides Nucleic Acids 23: 953-968.
17.
Ryabinin VA, Boutorine AS, Helene C, Pyshnyi DV,
Sinyakov AN (2004) Oligonucleotide-minor groove binder conjugates and their
complexes with complementary DNA: Effect of conjugate structural factors on the
thermal stability of duplexes. Nucleosides Nucleotides Nucleic Acids 23:
789-803.
QUICK LINKS
- SUBMIT MANUSCRIPT
- RECOMMEND THE JOURNAL
-
SUBSCRIBE FOR ALERTS
RELATED JOURNALS
- Journal of Alcoholism Clinical Research
- Dermatology Clinics and Research (ISSN:2380-5609)
- International Journal of AIDS (ISSN: 2644-3023)
- Journal of Cardiology and Diagnostics Research (ISSN:2639-4634)
- Journal of Forensic Research and Criminal Investigation (ISSN: 2640-0846)
- International Journal of Clinical Case Studies and Reports (ISSN:2641-5771)
- Journal of Immunology Research and Therapy (ISSN:2472-727X)