3936
Views & Citations2936
Likes & Shares
Many papers highlighted the phytochemicals anticancer action of N.sativa, but the peptides of N.sativa as anticancer option has not yet been investigated and, thus, this is the goal of this study.
MATERIALS AND METHODS
Methodology of current study
The current study employed many sequential databases and webservers to assess the anticancer bioactivity of N.sativa peptides as illustrated in Figure 1.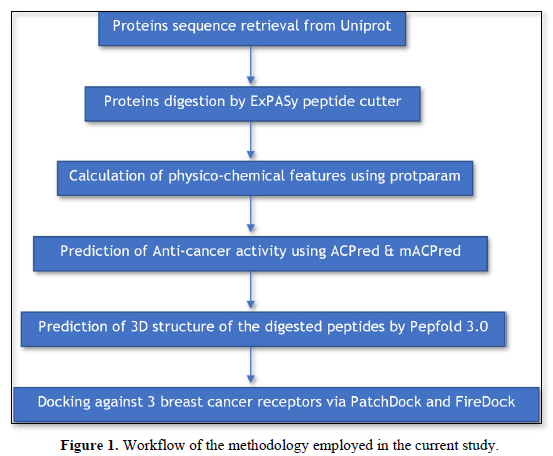
Retrieval of protein sequences
A total of 6 proteins served as precursors for the sought peptides: Thionin NsW1 (Uniprot ID# C0HJH9, 35 AA), Defensin D1 (Uniprot ID# P86972, 50 AA), Defensin D3r (Uniprot ID# A0A173AE14, 79 AA), CYC-like protein 1 (Uniprot ID# A0A166XVF4, 112 AA), Nigellin 1.1 (Uniprot ID# A0A1S4NYD1, 38 AA) and PsbA (Uniprot ID# A0A2U8T5R6, 23 AA). All of the protein sequences were retrieved from Uniprot database [19] using the corresponding ID.
Proteolysis
The selected proteins were subjected to proteolytic cleavage by trypsin using the webserver ExPASy peptide cutter tool [20]. Only the peptides having 5 or more AA were selected for further analysis.
Physico-chemical properties of fragmented peptides
The Physical and chemical properties of the digested peptides were calculated via the online tool Protparam [21] which include molecular weight (MW), theoretical isoelectric point (pI), aliphatic index and grand average of hydropathicity (GRAVY).
Assessment of ACP bioactivity
Two online tools were utilized for prediction of anti-cancer activity of the digested peptides: mACPred [22] and ACPred [23]. The two servers use different algorithms and prediction methods so only the best mutual results will be considered for further analysis.
3D structure modeling and prediction
The top best peptides that gave highest score of anti-cancer activity were adapted to predict the 3D structure de novo using Pepfold 3.0 server [24].
Docking
Given that the fragmented peptides have anti-cancer activity, 3 cancer receptors were chosen for as targets for molecular docking in order to confirm the anti-cancer activity of the produced peptides. PatchDock [25]was used as a platform for peptide-protein docking. Results of PatchDock were refined using FireDock server [26] to give the best 10 solutions along with their global energy and its details.
Visualization of docking interaction
Of the docked models, only the models giving the highest global energy got their peptide-protein interaction visualized via Discovery Studio 2021 software.
RESULTS AND DISCUSSION
To treat cancer efficiently, chemotherapeutic agents must reach the cancer microenvironment at high concentrations with no or less accumulation in off-target tissues [27]. In a variety of means, ACP can control cancer progression. Some can promote or block certain cancer receptors or signaling pathways while others serve as vehicles to carry therapeutics inside cancer cells or tissues [28]. Also, compared to proteins or antibodies, ACP is easier and cheaper to produce and design [29]. This glorifies the importance of ACP in the current research. So the aim of this paper is to produce peptides from N.sativa proteins and testing their anticancer activities.
Physico-chemical properties of the fragmented peptides
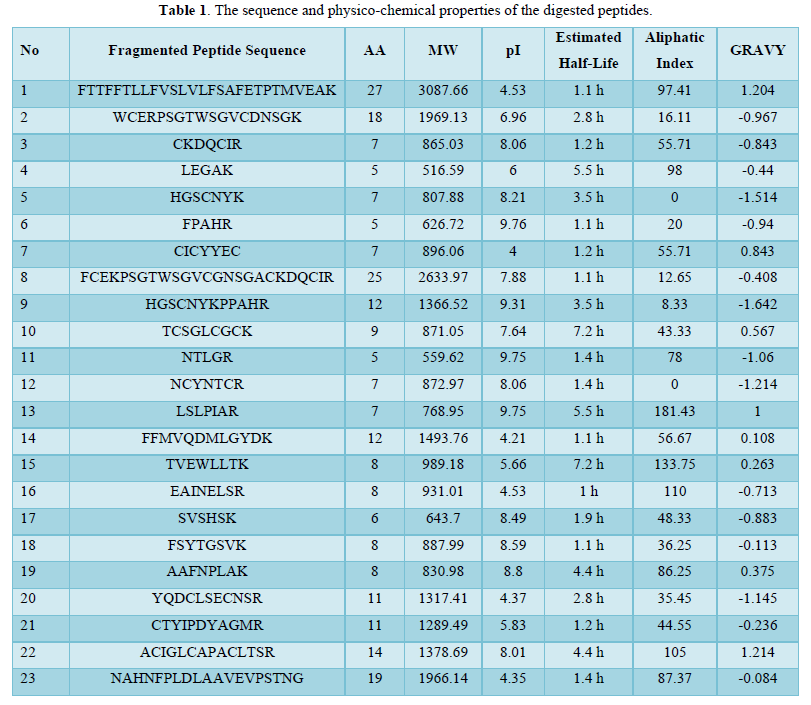
A set of 23 peptides were cleaved from 6 precursor proteins using trypsin. The amino acid range of the generated peptides was between 5-27. The theoretical pI values of most of the digested peptides were alkalic (>7) suggesting the abundance in basic AA of resulting peptides. The half-life was estimated in vitro of mammalian reticulocytes and most of the peptides had a half-life of 1-2 h. The rest of the characteristics are shown in Table 1.
Assessment of ACP bioactivity
Table 2 summarizes the ACP of the fragmented peptides based on the two tools mACPred and ACPred.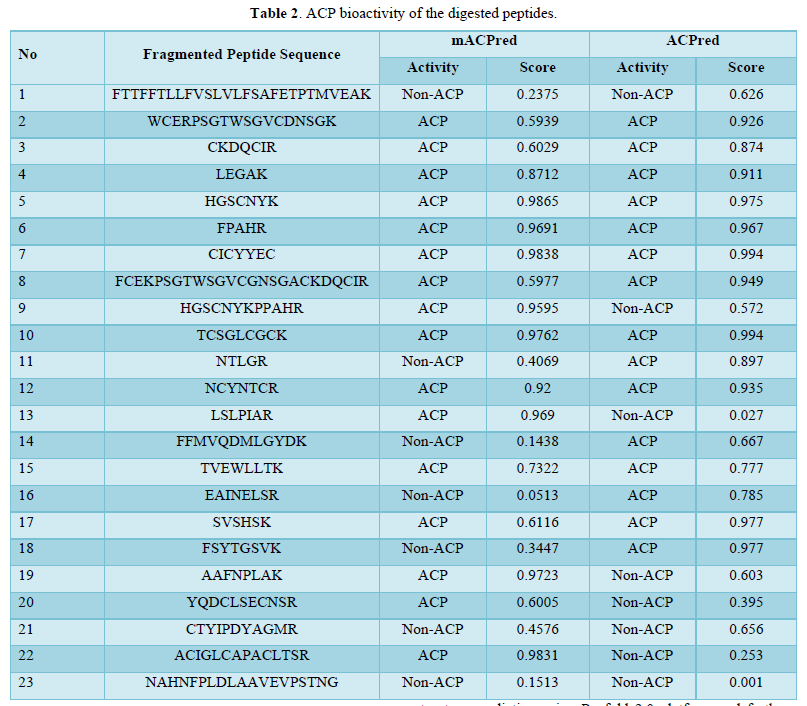
Of the 23 digested peptides, 16 exhibited anticancer activity estimated via mACPred tool. Similarly, 15 peptides exhibited anticancer activity in ACPred server. Among those, there were 11 mutual peptides with anticancer activity. It should be noted that these findings were extracted from only 6 proteins suggesting the effectiveness of N.sativa peptides against cancer.
3D structure modeling and docking
The top 3 (peptide 5, peptide 7 and peptide 10) peptides with mutual high score of anticancer activity were selected for 3D structure prediction using Pepfold 3.0 platform and further for docking studies. Docking studies were performed via PatchDock and the further refinement of docked poses through FireDock platforms. 3 breast cancer receptors were chosen as target for docking: epidermal growth factor receptor (EGFR; PDB ID#4HJO) estrogen receptor-α (ER-α; PDB ID# 3ERT) and matrix metalloproteinase-3 (MMP-3; PDB ID# 2D1O). ER-α activation is closely linked to the various stages of breast cancer [30]. Likewise, inappropriate activation of EGFR drives the tumorigenesis of lung and breast, among others [31]. With regard to MMP-3, overexpression of this extracellular enzyme has been correlated with many cancer types including breast cancer especially in the metastasis stage [32]. Results of docking are elucidated in Table 3.
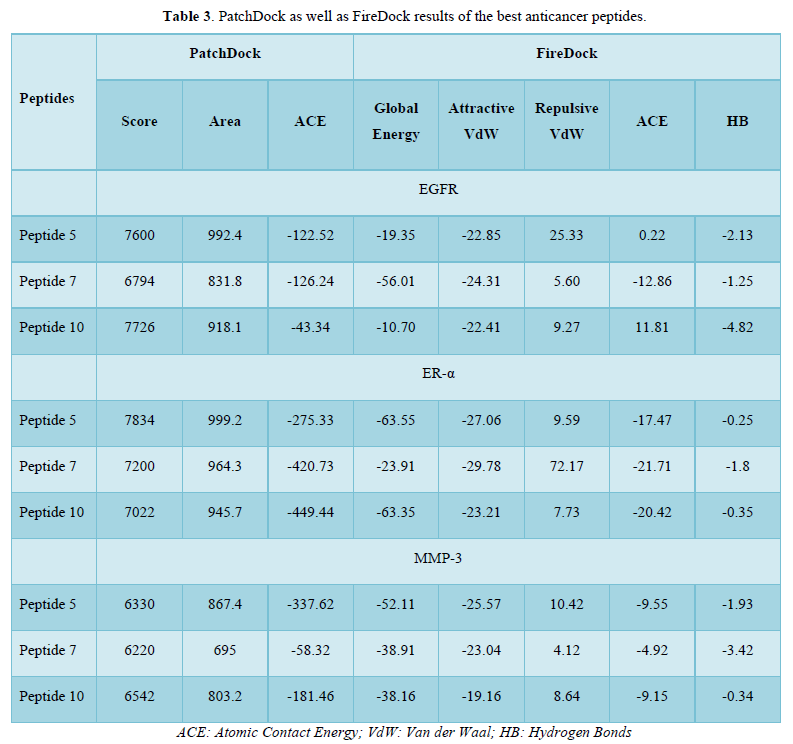
As shown in Table 3, peptide 7 greatly blocks EGFR with a global energy of -56.01 kcal/mole while the remaining peptides are not good enough for EGFR inhibition. With respect to ER-α, peptide 5 as well as peptide 10 are superior inhibitors with a global energy -63.55 and -63.35 kcal/mole. Of the top 3 peptides, peptide 5 are good candidate for inhibition of MMP-3 (global energy -52.11 kcal/mole) whilst peptides 7 and 10 had a global energy of -38.91 and -38.16 kcal/mole. Collectively, peptide 7 is a good candidate for blockade of EGFR whereas peptide 5 can be used as inhibitor of both ER-α and, along with peptide 10, MMP-3. Peptide-protein interactions of best poses are shown in Figure 2. These data suggest that the studied peptides fragmented from N.sativa can be utilized as ACP against breast cancer receptors. Surprisingly, peptide 7 and peptide 5 are even better than the reference inhibitors of EGFR and ER-α (Erlotinib and TAM) in terms of global energy estimated by FireDock (-50.90 and -60.2 kcal/mole; data not shown).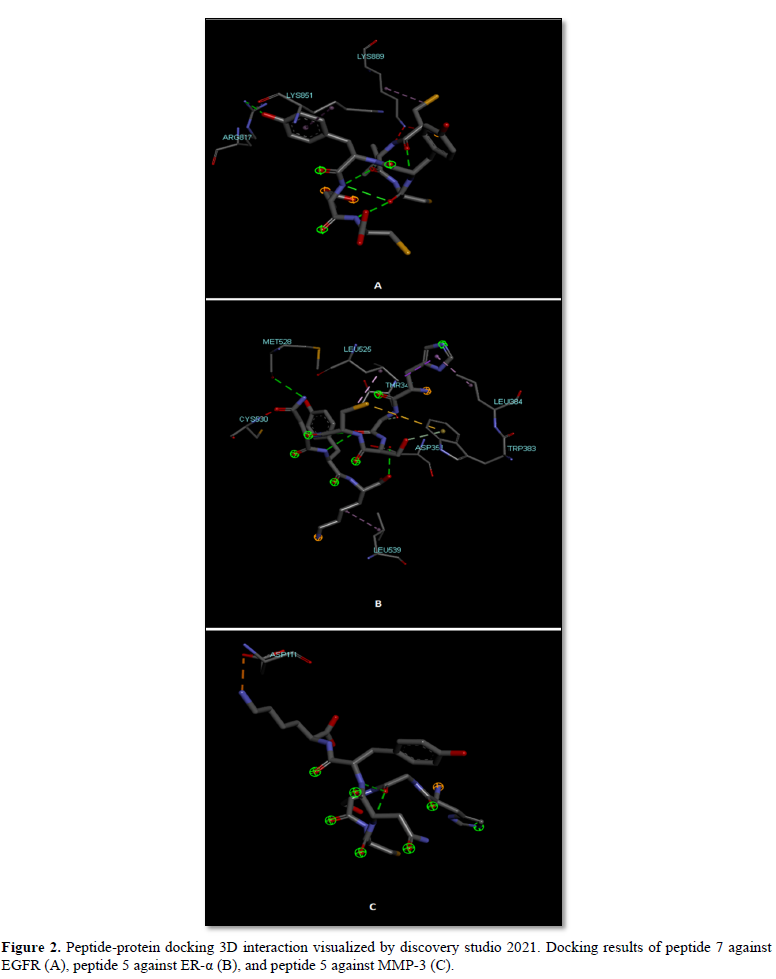
EGFR interacted with peptide 7 through VdW interactions imposed by two lysine residues within the active site pocket and one arginine residue through H-bonds. Similarly, Asp 351 and Met 528 formed H-bonds with peptide 5 and the rest interacting residues interacted via VdW forces with ER-α. Regarding MMP-3, peptide 5 formed electrostatic attractions with Asp 111 (Figure 2).
Accordingly, the present work demonstrates in-silico the anti-cancer activity of the peptides fragmented from some N.sativa proteins as predicted by ACPred and mACPred webservers and then validated by peptide-protein docking through PatchDock-FireDock platform. This candidates N.sativa as a superior source as therapeutic nutraceutical option against breast cancer theoretically.
CONCLUSION
In conclusion, 6 N.sativa proteins after proteolysis using trypsin enzyme gave rise to 23 peptides for which physico-chemical properties were calculated. 11 of which had ACP bioactivity as predicted by mACPred and ACPred platforms. Among them, peptides 5 (HGSCNYK), peptide 7 (CICYYEC), and peptide 10 (TCSGLCGCK) were the best in terms of the possibility score as ACP against 3 types of breast cancer receptors, EGFR, ER-α and MMP-3 as demonstrated by PatchDock and FireDock servers which necessitate in vitro assays confirmation. However, further analysis of the top peptides against wide array of other cancer types should be addressed. Also, the peptides with low anti-cancer activity should be assessed for different bioactivities.
- Khan MA, Chen H, Tania M, Zhang D (2011) Anticancer activities of Nigella sativa (black cumin). Afr J Tradit Complement Altern Med 8: 226-232.
- Majdalawieh AF, Fayyad MW, Nasrallah GK (2017) Anti-cancer properties and mechanisms of action of thymoquinone, the major active ingredient of Nigella sativa. Crit Rev Food Sci Nutr 57: 3911-3928.
- Ahmad A, Husain A, Mujeeb M, Khan SA, Najmi AK, et al. (2013) A review on therapeutic potential of Nigella sativa: A miracle herb. Asian Pac J Trop Biomed 3: 337-352.
- Butt MS, Sultan MT (2010) Nigella sativa: Reduces the risk of various maladies. Crit Rev Food Sci Nutr 50: 654-665.
- Gali-Muhtasib H, Roessner A, Schneider-Stock R (2006) Thymoquinone: A promising anti-cancer drug from natural sources. Int J Biochem Cell Biol 38: 1249-1253.
- Nickavar B, Mojab F, Javidnia K, Amoli MAR (2003) Chemical composition of the fixed and volatile oils of Nigella sativa from Iran. Zeitschrift Für Naturforschung C 58: 629-631.
- Salem ML (2005) Immunomodulatory and therapeutic properties of the Nigella sativa seed. Int Immunopharmacol 5: 1749-1770.
- Majdalawieh AF, Fayyad MW (2016) Recent advances on the anti-cancer properties of Nigella sativa, a widely used food additive. J Ayurveda Integr Med 7: 173-180.
- Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, et al. (2021) Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 71: 209-249.
- Rizvi M, Alam MS, Mehdi SJ, Ali A, Batra S (2012) Allelic loss of 10q23. 3, the PTEN gene locus in cervical carcinoma from Northern Indian population. Pathol Oncol Res 18: 309-313.
- Rahmani AH, Alzohairy M, Babiker AYY, Khan AA, Aly SM, et al. (2013) Implication of androgen receptor in urinary bladder cancer: A critical mini review. Int J Mol Epidemiol Genet 4: 150.
- Jo Y, Choi N, Kim K, Koo H-J, Choi J, et al. (2018) Chemoresistance of cancer cells: Requirements of tumor microenvironment-mimicking in vitro models in anti-cancer drug development. Theranostics 8: 5259.
- Lee Y-W, Chen T-L, Shih Y-RV, Tsai C-L, Chang C-C, et al. (2014) Adjunctive traditional Chinese medicine therapy improves survival in patients with advanced breast cancer: A population-based study. Cancer 120: 1338-1344.
- Rahmani AH, Alzohairy MA, Khan MA, Aly SM (2014) Therapeutic implications of black seed and its constituent thymoquinone in the prevention of cancer through inactivation and activation of molecular pathways. Evid Based Complement Alternat Med 2014: 724658.
- Tyagi A, Tuknait A, Anand P, Gupta S, Sharma M, et al. (2015) CancerPPD: A database of anticancer peptides and proteins. Nucleic Acids Res 43: D837-D843.
- Thundimadathil J (2012) Cancer treatment using peptides: Current therapies and future prospects. J Amino Acids 2012: 967347.
- Karami M, Chaleshgar M, Salari N, Akbari H, Mohammadi M (2022) Global Prevalence of Anemia in Pregnant Women: A Comprehensive Systematic Review and Meta-Analysis. Matern Child Health J 26(7): 1473-1478.
- Chiangjong W, Chutipongtanate S, Hongeng S (2020) Anticancer peptide: Physicochemical property, functional aspect and trend in clinical application (Review). Int J Oncol 57: 678-696.
- Consortium U (2019) UniProt: A worldwide hub of protein knowledge. Nucleic Acids Res 47: D506-515.
- Gasteiger E, Hoogland C, Gattiker A, Wilkins MR, Appel RD, et al. (2005) Protein identification and analysis tools on the ExPASy server. The Proteomics Protocols Handbook 2005: 571-607.
- Garg VK, Avashthi H, Tiwari A, Jain PA, Ramkete PW, et al. MFPPI–multi FASTA ProtParam interface. Bioinformation 12: 74.
- Boopathi V, Subramaniyam S, Malik A, Lee G, Manavalan B, et al. (2019) mACPpred: A support vector machine-based meta-predictor for identification of anticancer peptides. Int J Mol Sci 20: 1964.
- Schaduangrat N, Nantasenamat C, Prachayasittikul V, Shoombuatong W (2019) ACPred: A computational tool for the prediction and analysis of anticancer peptides. Molecules 24: 1973.
- Lamiable A, Thévenet P, Rey J, Vavrusa M, Derreumaux P, et al. (2016) PEP-FOLD3: Faster de novo structure prediction for linear peptides in solution and in complex. Nucleic Acids Res 44: W449-W454.
- Schneidman-Duhovny D, Inbar Y, Nussinov R, Wolfson HJ (2005) PatchDock and SymmDock: Servers for rigid and symmetric docking. Nucleic Acids Res 33: W363-W367.
- Andrusier N, Nussinov R, Wolfson HJ (2007) FireDock: Fast interaction refinement in molecular docking. Proteins 69: 139-159.
- Sutradhar KB, Amin M (2014) Nanotechnology in cancer drug delivery and selective targeting. Int Sch Res Notices 939378: 1-12.
- Hayashi MA, Ducancel F, Konno K (2012) Natural peptides with potential applications in drug development, diagnosis, and/or biotechnology. Int J Pept 2012: 757838.
- Grisoni F, Neuhaus CS, Gabernet G, Müller AT, Hiss JA, et al. (2018) Designing anticancer peptides by constructive machine learning. ChemMedChem 13: 1300-1302.
- Liu Y, Ma H, Yao J (2020) ERα, A Key Target for Cancer Therapy: A Review. Onco Targets Ther 13: 2183-2191.
- Sigismund S, Avanzato D, Lanzetti L (2018) Emerging functions of the EGFR in cancer. Mol Oncol 12: 3-20.
- Suhaimi SA, Chan SC, Rosli R (2020) Matrix Metallopeptidase 3 Polymorphisms: Emerging genetic Markers in Human Breast Cancer Metastasis. J Breast Cancer 23: 1-9.
QUICK LINKS
- SUBMIT MANUSCRIPT
- RECOMMEND THE JOURNAL
-
SUBSCRIBE FOR ALERTS
RELATED JOURNALS
- Journal of Agriculture and Forest Meteorology Research (ISSN:2642-0449)
- Journal of Astronomy and Space Research
- Advances in Nanomedicine and Nanotechnology Research (ISSN: 2688-5476)
- Food and Nutrition-Current Research (ISSN:2638-1095)
- Journal of Biochemistry and Molecular Medicine (ISSN:2641-6948)
- Journal of Veterinary and Marine Sciences (ISSN: 2689-7830)
- Journal of Genomic Medicine and Pharmacogenomics (ISSN:2474-4670)




