1134
Views & Citations134
Likes & Shares
Staphylococcus aureus is a serious human pathogen that causes vast range of contagious conditions both in hospitals and community settings. Methicillin resistant S. aureus (MRSA) are oftenly multidrug resistant in hospital and community that resulted in significant mortality and morbidity. Today, the methicillin resistant S. aureus (MRSA) had become endemic in a hospital worldwide. So, the need to quick diagnosis and identify of (MRSA) by using PCR technique. A total of 150 clinical specimens include: wound swabs, sputum, throat swabs, nasal swabs, pus and urine, that collected randomly from patients suffering of UTI, wound infection and upper respiratory tract infection, who attended the outpatients and inpatients Clinics of Alsadr Teaching Hospital and Al-Shefa General Hospital in Basrah City south of Iraq country. The identification of the methicillin resistant S. aureus (MRSA) from clinical specimens were performed depending on morphological and biochemical assay based positive cultures of S. aureus from clinical samples. Out of 150 clinical specimens, 61(40.66 %) of Staphylococcus aureus were isolated and identified by conventional methods, 20 (62.5 %) of these bacterial isolates were from ear pus samples followed by 18 (60.0 %) were from sputum, 11 (26.82 %) were from urine, 6 (28.57 %) were from nasal, 4 (21.05 %) were from throat and 2 (28.57 %) were from wound samples respectively. Also, 56 (37.33 %) of (CONS) were detected from clinical samples and they included Staphylococcus epidermidis which was frequently isolated bacterial species 41 (73.21%) followed by 15 (26.78 %) of Staphylococcus saprophyticus. All isolates of S. aureus were be examined for methicillin resistance by disc diffusion assay of oxacillin and cefoxitin antibiotics and by using modified Kirby-Bauer method. Inhibition zones according to the Clinical and Laboratory Standards Institute (CLSI, 2021) which recorded a higher resistance (100 %) to both antibiotics. PCR based molecular method was used for accurate identification of specific 16S rRNA gene and mec A gene in S. aureus (MRSA) isolates from clinical samples. The results of PCR amplification of 16srRNA specific gene from S. aureus isolates revealed that 52/61 isolates from a total 61 of S. aureus that detected in clinical samples through conventional methods with percentage of (85.24 %) had a clear band of approximately 228 bp which corresponds to identification of S. aureus strains. While, the PCR amplification of 42/52 isolates with percentage of (80.76 %) were generated a clear band of approximately 310 bp which corresponding to detect mec A gene in methicillin resistant strains of S. aureus isolates.
Keywords: Staphylococcus aureus, MRSA, Antibiotic resistance, Mec A gene, 16srRNAINTRODUCTION
Staphylococcus aureus is a Gram-positive bacterium and commensal microorganism that colonizes about 30 % of hygienic individuals from various body parts [1]. Its play a considerable role in causing of infections in both community and hospital acquired, that ranging from simple to life threating infections. Also, the S. aureus is regarded as a causative agent of a broad range of infectious diseases like the skin infections, endocarditis, osteomyelitis, bacteremia, necrotizing pneumonia, toxic shock syndrome, infections associated with foreign bodies, post-operative wound infections and food poisoning [2-4]. The infections that are causes by S. aureus due to owing a virulent gene which encodes variant of virulence factors like enzymes and toxins, among others. Furthermore, the virulence factor of S. aureus has rising with presence a strain that have antibiotics resistance as methicillin resistant S. aureus (MRSA) and vancomycin resistance S. aureus (VRSA) [5]. It persists to be one of the prevalent pathogens confronted in clinical practice additionally, it can cause intrinsic mortality and morbidity value. S. aureus can rapidly accommodate to the selective pressure of antibiotics that has consequence in the emergence and spreading of the methicillin- resistant S. aureus (MRSA) [6]. Methicillin- resistant S. aureus (MRSA) is a bacterial strain that resistant to the Beta- lactams, which comprise the penicillin’s and cephalosporins. Also, the antimicrobial resistance in the (MRSA) strains of S. aureus bacterium is correlated with the acquisition of a mobile genetic elements that called staphylococcal cassette chromosome mec, which carries the mec A gene that encoding the low- affinity of penicillin- binding protein 2 a and affords resistance to a B-lactam antibiotic [7,8].
The growing of drug-resistant virulent strains of Staphylococcus aureus particularly, (MRSA) is a critical problem in controlling and treatment of staphylococcal infections [9]. Further, due to the outbreak of infectious diseases that caused by various bacterial pathogens and development of resistance to variant antibiotics within it MRSA so, we need to detect and quick diagnose of these MRSA isolates from clinical samples to control and treated the infections related with this dangerous bacteria. Many of PCR based methods were progressed as an alternative approach for precise identification [10]. Amplification of specific 16SrRNA gene sequences (228 bp) is the most generally method that used for identification of S. aureus bacteria [11] and amplify the mec A gene sequences (310 bp) is used for detect the MRSA isolates in clinical samples [12].
The aim of this study is to detect and identification of specific 16S rRNA gene and mec A gene in (MRSA) isolates from clinical samples by PCR technique.
MATERIALS AND METHODS
Samples Collection
A total of 150 clinical samples include: wound swabs, sputum, throat swabs, nasal swabs, pus and urine, that will be collected randomly from patients suffering of UTI, wound infection and upper respiratory tract infection, who attended the outpatients and inpatients Clinics of Alsadr Teaching Hospital and Al-Shefa General Hospital during the period from 1st of January, 2022 to July, 2022.
All samples will be collected in sterile conditions with sterile containers then will be transmitted to the microbiology Lab. In the College of Medicine, University of Basrah for isolation and identification of associated pathogens and for detection the methicillin resistant Staphylococcus aureus (MRSA) by different cultivation and fundamental identification methods [13-15].
Samples processing
Bacterial isolation and identification
Apply the sterile conditions for equipment's related clinical samples. Each clinical sample were directly inoculated into plates of Mannitol Salt Agar (MSA) and Blood Agar, then incubated at 37 C0 for 24-48 h. then, all colonies from primary cultures were identified depending on the morphological features in culture media as beta hemolytic on Blood Agar and fermentation of the mannitol sugar on MSA. In addition to, performed of the biochemicals as diagnostic tests that comprise: catalase positive and coagulase positive also, the Gram staining that showed gram positive grape- like clusters. Finally, the subcultures will be done on Nutrient Agar plates for all isolates of S. aureus bacterium and other staphylococci when present.
Samples with isolation and outline of bacterial identification:
Clinical samples cultured on Blood agar and Mannitol salt agar, then incubated at 37C° for 24-48 h after that the subcultures was done on Nutrient agar for each isolate. Identification of isolates were confirmed by different biochemical tests [16,17].
Detection of MRSA strains (In vitro): by Perform of Antibiotic susceptibility test, all S. aureus isolates will be examined for methicillin resistance by disc diffusion assay of Oxacillin and Cefoxitin antibiotics. Staphylococcus aureus ATCC 25923 strain will be used as a positive control. The Multi Drug Resistant “S. aureus” MRSA isolates will be detected by their resistant to three or more antibiotic classes by using the modified Kirby- Bauer that following Clinical and Laboratory Standards Institute guidelines. -Most of clinical specimens confirmed by VITEK-2 compact system in Alsadr Teaching Hospital for detection of Multidrug Resistant (MRSA).
Molecular part:
Primers
The primers for confirm detection of bacterial species and detect the antibiotic resistance gene that were used throughout the present study are listed in Table 1.
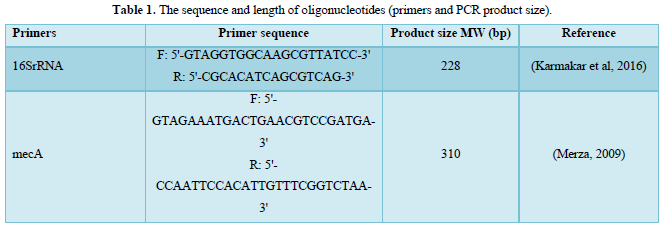
DNA Extraction and Purification:
Chromosomal DNA was extracted from the clinical isolates, one colony of each isolate cultured on a solid medium was inoculated into 5 ml of BHI (Brain Heart Infusion) and grown overnight at 37ºC. From these isolate cultures, the DNA was purified from bacterial cells by using Genomic DNA Mini kit which supplemented by the manufacturing company. Chromosomal DNAs obtained were used as templates for all PCR experiments. The PCR reactions were achieved in a Thermal Cycler. Before PCR assay, the DNA profile were carried out by using bacterial DNA and loading buffer without thermal cycling conditions, and according to the following Steps of DNA extraction
Step 1
- Cultured bacterial cells were transferred to 1.5 ml micro centrifuge tube, the sample was centrifuged at 14000xg for 1 min and the supernatant was discarded.
- An aliquot of 200 µl of Gram+ buffer (lysozyme buffer) were added to the sample and re-suspend the cell pellet by shaking vigorously or pipetting.
- The sample was incubated at 37 C for 30 min, during incubation, the tube was inverted every 10 min.
- Adding 20 µl of proteinase K to sample then mixed by vortex then incubate at 60 C° for 10 min, during incubation, invert the tube every 3 min.
- The lysis step was proceeded for the cultured cell protocol.
Step 2
- 200 µl of GB Buffer was added to the sample and mixed by vortex for 10 seconds incubated at 70°C for 10 min. During incubation, the tube was inverted every 3 min. and incubated the required Elution Buffer at 70°C (for step 5).
Step 3
- 200 µl of absolute ethanol was added to the sample and mixed by shaking vigorously. If precipitate appears, break it up by pipette.
- A GD Column was placed in a 2 ml collection tube.
- All of the mixture (including any precipitate) was transferred to the GD column, and then centrifuged at 14000xg for 2 min.
- Discarding the 2ml collection tube containing the flow-through then placed the GD column in a new 2 ml collection tube.
Step 4
- 400 µl of W1 Buffer was added to the GD column, and then Centrifuge at 14000xg for 30 sec.
- The flow-through discarded and the GD Column was placed back in the 2 ml Collection Tube.
- 600 µl of Wash Buffer (ethanol) was added to the GD Column, and then Centrifuged at 14000xg for 30 sec.
- The flow-through discarded again for 3min at 14000xg to dry column matrix.
Step 5
- The dried GD Column was transferred to a clean 1.5 ml micro centrifuge tube.
- 100 µl of pre-heated Elution Buffer or TE was added to the center of the column matrix.
- The sample was left to stand for 3-5 min or until the Elution Buffer or TE absorbed by the matrix.
- Centrifuged at 14000xg for 20 seconds to elute the purified DNA.
Amplification of the Genes of S. aureus Isolates
Amplification of the both genes (16SrRNA and mec A) were carried out by PCR thermo cycler instrument. The total volume of reaction mixture was 20 µl for amplification both genes (Table 2).
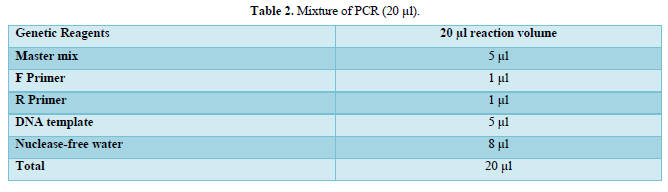
Confirmation of S. aureus isolates were done by using the primer of 16SrRNA specific gene while, the other primer of mec A gene used to detection the gene which responsible of resistant to methicillin.
PCR Thermo cycling conditions:
The PCR prepared tubes were placed in the machine of PCR and the correct PCR cycling program parameters conditions were installed as shown in Tables 3 & 4.
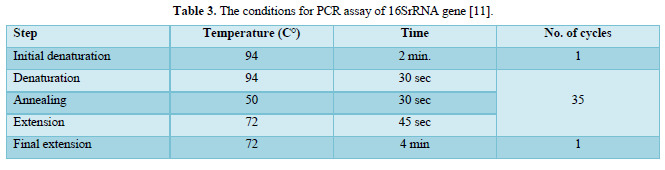
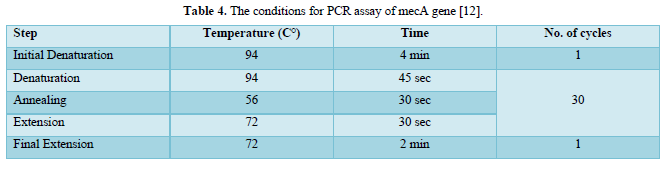
All requirements for technical and preparations of agarose gel electrophoresis for DNA detection and target gene analysis were implemented by Bartlett and Stirling.
Preparation of agarose gel and DNA loading [18]:
To separate DNA fragments, agarose gel in different concentrations were used (about 0.8 % for the extracted DNA and 1.2- 2 % for visual checking of the specific PCR products). Agarose powder is added to (100 ml) TBE buffer that previously prepared (90 ml D.W. was added to 10 ml TBE buffer 10 x, to obtain the final concentration was 1 x) to the desired concentration. Kindly, in equilibrated gel tray earlier set with combs fixed in the middle and end depending upon the samples that need to loading, also, the ends of gel tray were sealed. The glass baker which contain the agarose powder in TBE buffer was put in microwave to dissolved the agarose powder within TBE buffer and added in Carefully form the ethidium bromide in concentration of 0.5 mg/ml. The prepared gel was poured to cool and solidify at the room temperature waiting 30 min. and still in its plastic tray after that, the combs and seal were removed gently from the casting tray, the combs made wells that used for loading DNA samples.
The samples containing DNA mixed with 1/10 of loading buffer then, pipetted into the wells. Lid was placed on the apparatus and the power supply, then, the current was applied. Generally, the gel was run at 5 v/cm for about 30 min to 1.5 h to separate the DNA fragments for detect of presence of DNA in samples and for PCR products respectively. DNA bands were visualized by U.V. trans illuminator at 365 nm wave length documentation system, then photographed by digital or phone camera.
Detection of specific genes by polymerase chain reaction:
The detection of S. aureus specific species gene, in addition to the genes that encoding the resistance for oxacillin and cefoxitin antibiotics were done by amplification of specific sequences within the target gene by using the polymerase chain reaction technique. The experiment was performed by using the mixture of a specific sets of primers designated for each targets genes that were mixed with the DNA samples (as a template) and master mix reagent which contain (Taq polymerase, PCR buffer, MgCl2 and dNTPs), the final constituents was the nuclease free water, the reaction mixtures were mixed for each targets genes then, transfer to thermal cycler machine to start the reaction depending on the steps of specific program [19,20].
Detected of S. aureus isolates from clinical samples by conventional methods
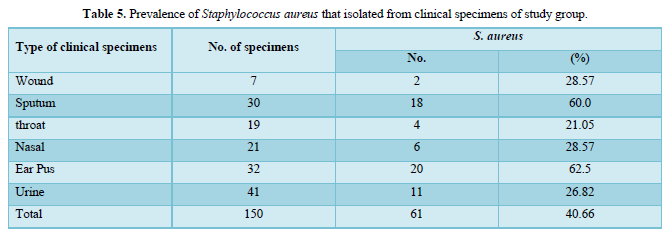
Out of 150 clinical specimens that were included in this study, a total of 61 Staphylococcus aureus were isolated and identified. Twenty (20) of these bacterial isolates were from ear pus samples will percent of (62.5 %) followed by eighteen (18) were from sputum in percent (60 %) then, eleven (11) were from urine with (26.82 %), six (6) were from nasal with (28.57 %), four (4) were from throat and two (2) were from wound samples, with percent of (21.05 %) and (28.57 %) respectively as shown in Table 5.
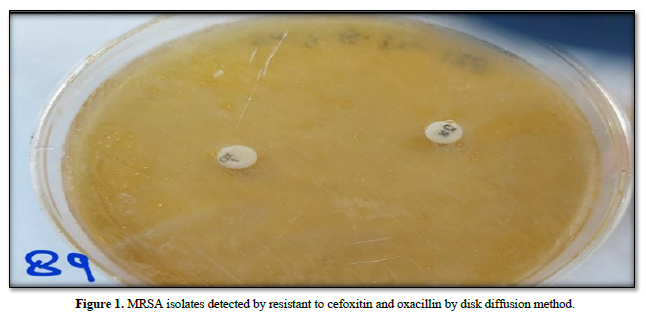
Identification of MRSA isolates by using the modified Kirby-Bauer method according to (CLSI, 2021)
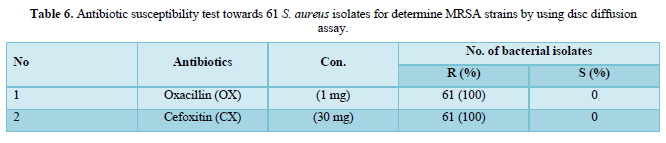
Out of 150 samples, 61 (40.66 %) were found to be infected with S. aureus isolates and the results of (61) isolates of S. aureus against cefoxitin (30 μg) and oxacillin (1 μg) antibiotic disks by disk diffusion assay showed that, all bacterial isolates were resistant to both antibiotic disks in rate (100 %) and were considered as (MRSA), (Table 6 & Figure 1).
Frequency of coagulase negative (CONS) Staphylococcus spp. from clinical specimens:
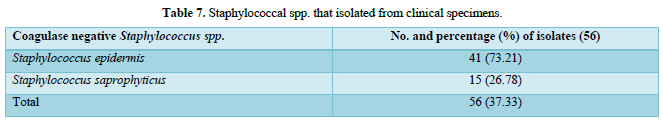
Out of 150 specimens, the coagulase negative (CONS) Staphylococcus spp. were isolated from patients in 56 (37.33 %) and they consisted of Staphylococcus epidermidis which was appeared as the high frequently isolated bacterial species 41 (73.21%) followed by Staphylococcus saprophyticus 15 (26.78 %) as appeared in Table 7.
Molecular part:
Detection of 16S rRNA and mec A genes from S. aureus isolates in different clinical specimens:
The purified fragments DNA of 61 S. aureus isolates from different sites of infection in human were conducted for PCR assay to detect the presence of 16srRNA, and mec A genes as shown in Table 8. The results of PCR amplification of 16srRNA gene from S. aureus isolates revealed that 52/61(85.24 %) isolates had a clear band of 228 bp which corresponds to identification of S. aureus strains. While, PCR amplification of 42/52(80.76 %) isolates were generated a clear band of 310 bp which corresponding to detect mec A gene in methicillin resistant strains of S. aureus isolates, as appeared in Figures 2-4.
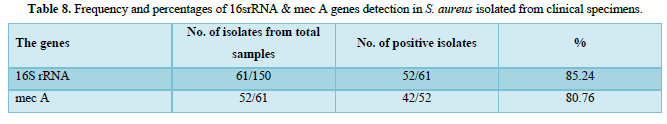

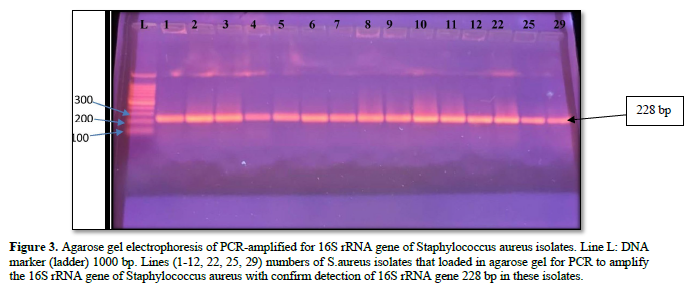
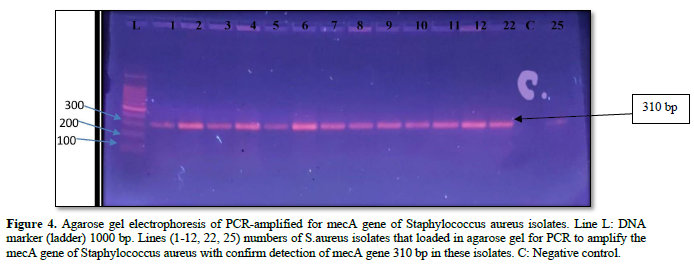
Detection of S. aureus isolates by molecular and conventional methods:
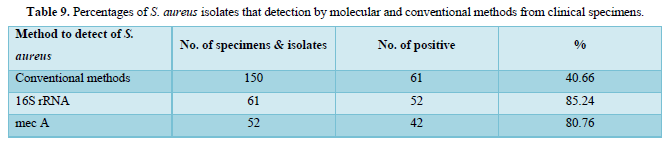
PCR assay was used to detect the presence of 16srRNA specific for S. aureus and mec A gene in bacterial isolates through the amplification of 16srRNA gene and mec A gene from S. aureus isolates as shown in Table 9. The results were revealed that 52/61 (85.24 %) isolates had a clear band of approximately 228 bp which corresponds to specific identification of 16srRNA gene in S. aureus isolates, and 42/52 (80.76 %) isolates were generated a clear band of approximately 310 bp which corresponding to detect mec A gene in methicillin resistant strains of S. aureus isolates. While, in conventional methods which that be used for detection of S. aureus and methicillin resistant strains from different clinical specimens were revealed, out of 150 tested samples, 61 (40.66 %) were found to be infected with S. aureus isolates and all isolates of S. aureus were (100 %) resistant to oxacillin. Cefoxitin, ampicillin, methicillin and amoxicillin respectively that previous mentioned with details in Tables 5 & 6.
In this study the identification of the S. aureus and other Staphylococcus spp. from clinical samples were performed depending on morphological and biochemical assay including gram stain, catalase test, coagulase test, changes on mannitol salt agar and blood agar media in addition to susceptibility against Novobiocin disc. This results that were obtained from the traditional methods for isolation and identification of S. aureus isolates from clinical samples agreed with the results of Oladipo [21]. Also, from 150 clinical specimens that were implicated in current study, a total of 56 other Staphylococcal spp. (CONS) that were identified and represented the rate of (73.21%) in 41 isolates of Staphylococcus epidermis, followed by S. saprophyticus with rate of (26.78%) in 15 isolates. The results of current study were in congruence with the results of Mohammed [22] and Deyno [23] while the study of AL-Mosawi [24], show the low rates in prevalence of (CONS) which isolated from UTI patients in compare with rates of (COPS), these results were somewhat in accordance with our results.
The identification of the methicillin resistant S. aureus (MRSA) from clinical specimens were performed depending on morphological and biochemical assay based positive cultures of S. aureus from clinical samples. Out of 150 a total of 61 Staphylococcus aureus were isolated and identified. All isolates of S. aureus were examined to detect the methicillin resistance strains, that were perform by disc diffusion assay to Oxacillin and Cefoxitin antibiotics with the modified Kirby-Bauer method on Muller Hinton agar (MHA), the cultured plates of bacteria were examined after incubation period at 37°C for 24 h then by the following of Clinical and Laboratory Standard Institute were determined the zone diameter stop points of inhibition for the isolates of S. aureus to the closest millimeter (mm). shown that, the rates of resistance were highest and record (100%) for Oxacillin and Cefoxitin. In present study, the percentage of antibiotic resistance towards Oxacillin was (100%), that is accordance with the study of Ifra [25] who found that, the S. aureus was recorded in highly resistant towards Oxacillin in rate 100%. Also, the study of and Onemu [26] in university of Benin Hospital, Benin City which report the prevalence of MRSA in rate 79%, this finding was, to some extent, in agreement with the results of current study concerning with the isolation rate of MRSA. And in study of Lin [27] which was detected, the infectious cases by S. aureus (MRSA) particularly nosocomial infection in a Medical Center, that corresponding to 53 (66.3%) which represent the rate of MRSA prevalence, those finding were to some accordance with on the other hand, both studies were recorded a low percentages of MRSA isolates than detected in current study. Also, in study of detected the cefoxitin resistance 19 (73.07 %) of S. aureus isolates from patients with CSOM infections this result was constant with our results and out of 82 clinical specimens (31.70 %) percent of S. aureus isolates which detect low rat in compare with current study. In addition, other study shows the percentage rate of identification S. aureus isolates from patients with caries were, out of 50 clinical samples 26 (52 %) this results of were constant with present study.
In study of study was recorded the MRSA prevalence in rate (48.8%) which is comparable to that obtained by Olowe [28] who is reported the prevalence of MRSA in rate (47.8%) in Oshogbo, Ladoke Akintola University of Technology College of Health Sciences, South western Nigeria. Furthermore, in study of, the results of detected MRSA isolates from clinical specimens was recorded in rate of prevalence (24.75%) which is comparable to the results that reported by the study of Hujier [29] in Gaza, Palestine. While, in present study the rate of resistance in MRSA isolates are higher and were recorded (100%) for Oxacillin, Cefoxitin, Ampicillin, Amoxillin and Methicillin, these results were consistent with the results that reported in study of Aboud [30]. Our finding was disagreement with the results of that related with rates of sensitivity for Methicillin (51.2%) and Oxacillin (46.5%) and consistence with its results which related to other antibiotics used in this study.
The results of antibiotic susceptibility test and detection of mecA gene in study of Naorem [31] showed, 33 (94.28 %) strains were MRSA while 2 (5.72 %) strains were MSSA, this study agrees with present study and such a validation process was reported by other researchers [32-35]. The results of current study that related with PCR amplification of 16srRNA gene from S. aureus isolates revealed that 52 isolates in (85.24 %) percentage had a clear bands of approximately 228 bp which corresponds to identification of S. aureus strains, While, PCR amplification of 42 isolates in (80.76 %) percentage were generated a clear bands of approximately 310 bp which corresponding to detect mec A gene in methicillin resistant strains of S. aureus isolates, The present study is constant with the study of Al-Ashmawy [36] in which most of recovered S. aureus isolates were genetically verified as MRSA strains by molecular detection of the mecA gene. Further, in study of the percentage (90.7 %) of S. aureus isolates which identified by conventional methods were confirmed by amplification of the 16S rRNA gene, the current study agreed of those finding. The results of current study were somewhat in agreement with the results of Malihe [37] also, the high prevalence rates (57 %-70 %) of MRSA isolates in Alzoubi [38] study which documented among Jordanian hospitalized adults were coinciding with results of present study. All S. aureus isolates contained the resistance genes for (mecA) and penicillin were reveled in study of de which is consistent with results of this study. In study of Azimian [39] the results of PCR for mecA showed that 110 strains of S. aureus with percent (47 %) had mecA gene lower than we detected in present study but the study of Kot [40] which detect the methicillin resistant S. aureus (MRSA) in higher percentages about (80 %) by PCR amplification of mecA gene this result is agreement with current study. Gadban [41] study showed the result of amplifying 16S rRNA was given positive for identify the S. aureus and MRSA strains in all isolates, the 18 (100 %) were pvl positive, results of present study were consistent with those finding. While, in conventional methods that used for detection of S. aureus and methicillin resistant strains from different clinical specimens in present study were revealed, out of 150 samples, 61 (40.66 %) were found to be infected with S. aureus and all isolates of S. aureus were (100 %) resistant to oxacillin, Cefoxitin, ampicillin, methicillin and amoxillin respectively. Kader [42] study reported that, (88.24 %) of S. aureus isolates were resistant to Oxacillin and methicillin discs, and the current results were in constant with those results. Also, in study of showed the highest percentage of MRSA strains were isolated from respiratory tract samples (49%) followed by urine cultures (43%) and nasal swabs (35%), the results of present study were somewhat in consistent with these results. In addition, the current study is along with the studies of whose found that, the coagulase positive S. aureus isolates were recorded a highly resistant against oxacillin antibiotic (100 %). Furthermore, study detected out of 150 swab samples of diabetic foot ulcer only 21 isolates which include 18 (85.7 %) was identified as S. aureus, the other 3 (14.3 %) isolate was identified as Staphylococcus spp. and the results of vitek *2 showed all 18 (100 %) S. aureus isolates were resistance to oxacillin and Cefoxitin discs, the results of current study were somewhat in agreement with those finding.
Many laboratories still favor using of Oxacillin in detection of MRSA strains, that because of the Oxacillin antibiotic preserve its activity through the storage best than the Methicillin antibiotic also, more likely to detect hetero resistant strains [43]. In hospital and according to the doctors, there are many critical cases that were infected with S. aureus that are treated with glycopeptides (as Vancomycin or Teicoplanin) which may lead to create of a new strain of resistant bacteria to these mention antibiotics. In a particular concern of MRSA strains that are starting to develop the resistance to Vancomycin antibiotic, which is at present the most effective of antibiotic inverse MRSA. In current study many factors may have participated in the above levels that mentioned of resistance towards the tested of antibacterial drugs which selected for study the susceptibility levels to S. aureus isolates including, the antibiotics that are misused by the health occupational, un skilled practitioners and a lay person. The incidence of MRSA in present study could be imputed to many agents, despite of the Methicillin not being routinely used against S. aureus infections. Additional to antibiotic stress, the horizontal gene transfer is considered as a contributing agent in the incidence of antibiotic resistance in clinical isolates. Then, it has been proposed that a high prevalence in resistance to a particular antibiotic dose not constantly reflected in antibiotic consumption [44,45]. Another contributed agent by the using of antimicrobials in food of animals. Antibiotics are usually added to feed for enhance the growth in animals, in particular the dairy sheep, cattle and poultry [46,47]. Recurrent traveling is a supplementary factor for transmitting the resistance in bacterial strains between countries, the misusing of antimicrobial is another participate factor [48]. In Iraq, there is a current practice that antibiotics can be purchased in absence of formula which leads to misusing of antibiotic drugs by a public thus, participating to the emanation and diffusion of antimicrobial resistance. Other causal factors involve a condition that relating to the public health as poor hospital hygienic accounting for the spreading of resistant bacteria and inadequate surveillance [49]. In present study, the prevalence of MRSA isolates were high as in studies of these studies in Iraq country thus, compulsory institute guidelines for detection of MRSA and controlling in the hospital by the daily oversight of a clinical laboratories for MRSA isolates, approach of monthly predictable cultures that monitoring of inpatients believed to be at high risk in conquest of MRSA infection also, inspection of hospital personnel, regular of hand washing and disinfecting by hospital personnel with policy regulation of antibiotics in usage and prescription, acquisition of a new and additional information routinely from antimicrobial susceptibility testing to bacterial isolates and monitor the testing of these bacterial isolates with surveillance the antibiotics resistance. All of which that mention above, are decisive for a good clinical practice and for logical polices against the resistance of antibiotics [50].
CONCLUSION
The results of current study that related with PCR amplification of 16srRNA gene and mec A gene in S. aureus isolates which previously detected by conventional methods showed, most S. aureus isolates had a clear band of 228 bp and 310 bp that identical to these genes in this bacterium with corresponds to identification of S. aureus (MRSA) strains. So, the PCR analysis effective in confirm detection the specific 16S rRNA gene of S. aureus isolates and mecA gene in methicillin resistant strains of these bacterial isolates.
Also, in current study, the higher resistance of MRSA isolates was illustrious to Oxacillin, Cefoxitin and in many other studies in Iraq country detected that so, some rules must follow to reduce the spreading of MRSA strains include, inspection to MRSA isolates among the reservoir and distributer of MRSA strains in hospitals (healthcare workers and patients). Also, the nomination of antimicrobial agent should be based on the in vitro susceptibility test with a hospital based antibiotic policies that must be robustly followed and firm surveillance of drug resistance to all bacterial pathogens are required in both hospital patients and the national level.
-
- Aboud, W. A. A (2019). Molecular Detection of Vancomycin and Methicillin-Resistant Genes of Staphylococcus aureus isolated from Animal and Human specimens. A thesis submitted to the council of the College of Veterinary Medicine in University of Basrah as requirement for the Degree of Master of Science in Veterinary Medicine/ Microbiology.
- Aboud, W. A. A., & Khudaier, B. Y. (2018). Molecular Detection of Methicillin-Resistant Staphylococcus aureus isolated from MILK and CHEESE of COW and BUFFALOES IN BASRAH CITY. Basrah Journal of Veterinary Research, 17(3).
- Ako-Nai A, Adeyemi F, Aboderin O, Kassim O. (2005). Antibiotic resistance profile of Staphylococci from clinical sources recovered from infants. Afr. J. Biotechnol, 4(8): 816-822.
- Al-Ashmawy, M. A., Sallam, K. I., Abd-Elghany, S. M., Elhadidy, M., & Tamura, T. (2016). Prevalence, molecular characterization, and antimicrobial susceptibility of methicillin-resistant Staphylococcus aureus isolated from milk and dairy products. Foodborne Pathogens and Disease, 13(3), 156-162.
- Ali MA, Abbasi SA, Arifs S, Mirza I A. (2007). Nosocomial infections due to Methicillin Resistant Staphylococcus aureus in hospitalized patients. Pak J Med Sci, Vol. 23 No.4.
- AL-Mosawi R M M (2010). Isolation and identification of resistance Staphylococcus aureus to vancomycin & Methicillin from outpatients clinic in AL-Shefa General Hospital in Basrah district. University of Thi-Qar Journal of Science, 2(3), 57-69.
- Al-Mosawi, R. M. (2018). Microbiological study and antimicrobial susceptibility pattern of ear infections in patients with Chronic Suppurative Otitis Media (CSOM) in Basrah Province. University of Thi-Qar Journal of Science, 2(4), 109-116.
- Al-Mosawi, R. M., Jasim, H. A., & Haddad, A. (2023). Study of the antibacterial effects of the starch-based zinc oxide nanoparticles on methicillin resistance Staphylococcus aureus isolates from different clinical specimens of patients from Basrah, Iraq. AIMS microbiology, 9(1), 90.
- Alzoubi, H., Aqel, A., & Abu-Helalah, M. (2013). Prevalence of methicillin resistant Staphylococcus aureus nasal carriage and its antibiogram in healthcare workers from South of Jordan. Journal of High Institute of Public Health, 43(1), 1-12.
- Appelbaum, P. C. (2006). MRSA—the tip of the iceberg. Clinical microbiology and infection, 12, 3-10.
- Azimian A, Najar-Pirayeh S., Mirab-Samiee S, Naderi M. (2012). Occurrence of methicillin resistant Staphylococcus aureus (MRSA) among clinical samples in tehran-iran and its correlation with polymorphism of specific accessory gene regulator (AGR) groups. Brazilian Journal of Microbiology, 43, 779-785.
- Bartleet JMS and Stirling D. (1998). PCR Protocols: Methods in Molecular Biology. 2 nd. Humana Press Inc. Totowa. NJ.
- Bitrus AA, Peter OM, Abbas MA, Goni MD. (2018). Staphylococcus aureus: A Review of Antimicrobial Resistance Mechanisms. Veternary Sciences: Research and Reviews. 4(2).
- Bitrus AA, Peter OM, Abbas MA, Goni MD. (2018). Staphylococcus aureus: A Review of Antimicrobial Resistance Mechanisms. Veterinary Sciences: Research and Reviews, 4 (2).
- Brown, D. F., Edwards, D. I., Hawkey, P. M., Morrison, D., Ridgway, G. L., Towner, K. J., & Wren, M. W. (2005). Guidelines for the laboratory diagnosis and susceptibility testing of methicillin-resistant Staphylococcus aureus (MRSA). Journal of antimicrobial chemotherapy, 56(6), 1000-1018.
- Cappuccino JG and Welsh CT. (2017). Microbiology: a laboratory manual, Pearson education.
- Clinical and Laboratory Standards Institute (CLSI) (2021). Performance standards for antimicrobial susceptibility testing, 31st Ed., Malvern.
- Collee JG, Fraser AG, Marmino BP, Simons A. (1996). Mackin and McCartney Practical Medical Micrbiology. 14 th ed. The Churchill Livingstone, Inc. USA.
- Collier L ., Balows A. and Sussman M. (1998). Topley and Wilsons Microbiology and Microbial Infections , Volume 2: Systemic Bacteriology , 9th Edition; 577-617.
- Collier L ., Balows A. and Sussman M. (1998). Topley and Wilsons Microbiology and Microbial Infections , Volume 3: Bacterial Infection , 9th Edition; 231-48.
- Danzmann, L., Gastmeier, P., Schwab, F. and Vonberg, R.P. (2013). Health care workers causing large nosocomial outbreaks: A systematic review. BMC Infectious Diseases, 13, p.98.
- Deyno S, Fekadu S and Seyfe S. (2018). Prevalence and antimicrobial resistance of coagulase negative staphylococci clinical isolates from Ethiopia: a meta-analysis. BMC microbiology, 18(1), 1-11.
- de Oliveira, T. L. R., Cavalcante, F. S., Chamon, R. C., Ferreira, R. B. R., & Dos Santos, K. R. N. (2019). Genetic mutations in the quinolone resistance-determining region are related to changes in the epidemiological profile of methicillin-resistant Staphylococcus aureus isolates. Journal of Global Antimicrobial Resistance, 19, 236-240.
- Emel B, Banu BB, Mehmet AB, Taner I, Erkan Y. (2010). Aplication of PCR-RFLP of gap gene method as a molecular typing tool for coagulase negative Staphylococci from bovine and human origin identified with VITEK 2. Afr J. Microbiol Res, 775-82.
- Feingold BJ, Silbergeld EK, Curriero FC, van Cleef BA, Heck ME and Kluytmans JA. (2012). Livestock density as risk factor for livestock-associated methicillin-resistant Staphylococcus aureus, the Netherlands. Emerging infectious diseases, 18(11), 1841.
- Forbes BA, Daniel FS, Alice SW. (2007). Bailey and Scott's diagnostic microbiology. 12 th. Ed. Mosby Elsevier company, USA.
- Gadban, T. H., Al-Amara, S. S., & Jasim, H. A. (2020). Screening the frequency of panton-valentine leukocidin (pvl) gene between methicillin resistant Staphylococcus aureus isolated from diabetic foot patients in Al-Basrah governorate, south of Iraq. Sys Rev Pharma, 11, 285-290.
- Gomes, A. R., Vinga, S., Zavolan, M., & De Lencastre, H. (2005). Analysis of the genetic variability of virulence-related loci in epidemic clones of methicillin-resistant Staphylococcus aureus. Antimicrobial agents and chemotherapy, 49(1), 366-379.
- Greenwood D, Slack RCB. and Peutherer JF. (2002). Medical Microbiology, 16 th Edition; Churchill Livingstone , UK.
- Hujier, N. A., & Sharif, F. A. (2008). Detection of methicillin-resistant Staphylococcus aureus in nosocomial infections in Gaza Strip. African Journal of Microbiology Research, 2(9), 235-41.
- Kader O, Ebid S, Mostafa Nancy, El Sayed S, Ghazal A (2011). Detection of Community Acquired Methicillin Resistance Staphylococcus aureus among Staphylococcus aureus isolates. J. Am. Sci. 7 (1): 1109.
- Kärki T, Napoli C, Riccardo F, Fabiani M, Dente M G, Carballo M, ... & Declich S. (2014). Screening for infectious diseases among newly arrived migrants in EU/EEA countries—varying practices but consensus on the utility of screening. International journal of environmental research and public health, 11(10), 11004-11014.
- Karmakar A, Dua P, Ghosh C. (2016). Biochemical and molecular analysis of Staphylococcus aureus clinical isolates from hospitalized patients, Hindawi Publishing Corporation. Can. J. Infect. Dis. Med. Microbiol.2016, 1-7.
- Katayama Y, Ito T and Hiramatsu K. (2000). A new class of genetic element, staphylococcus cassette chromosome mec, encodes methicillin resistance in Staphylococcus aureus. Antimicrobial agents and chemotherapy, Vol. 44 Issue 6 Pages 1549-1555.
- Kluytmans J, Belkum AV, Verbrugh H. (1997). Nasal carriage of Staphylococcus aureus; Epidemiology, underlying mechanisms and associated risk. Clin Microbiol Rev, Volume 505-520.
- Kot, B., Wierzchowska, K., Piechota, M., & Grużewska, A. (2020). Antimicrobial resistance patterns in methicillin-resistant Staphylococcus aureus from patients hospitalized during 2015–2017 in hospitals in Poland. Medical Principles and Practice, 29(1), 61-68.
- Kumurya, A. S. (2015). Detection of Staphylococcus aureus specific gene and simultaneous confirmation of methicillin resistant Staphylococcus aureus (MRSA) by polymerase chain reaction. Clin Med J, 1(3), 88-93. Lee PY, Costumbrado J, Hsu C, Kim YH. (2012).
- Agarose Gel Electrophoresis for the separation of DNA Fragments. Journal of Visualized Experiments, (62). Dio:10.3791/3923.
- Lin, D., Ou, Q., Lin, J., Peng, Y., & Yao, Z. (2017). A meta-analysis of the rates of Staphylococcus aureus and methicillin-resistant S aureus contamination on the surfaces of environmental objects that health care workers frequently touch. American journal of infection control, 45(4), 421-429.
- Macfaddin JF. (2000). Biochemical tests for identification of medical bacteria. 3 rd ed. Lippincott Williams and Wilkins USA.
- Malihe, H.; Hassan,M. and Mahboobeh, M. (2011). Detection of the antibiotic resistance genes in Staphylococcus aureus isolated from human infections and bovine mastitis. African Journal of Microbiology Research, 5(28), pp.5132- 5136.
- Merza N S. (2009). Application of Specific PCR assayes for detection of Methicillin-Resistant Staphylococcus aureus (MRSA). Sci. J.Univ.Zakho. 2(1), 65-73. (Change the reference number 12 from Othman et al., 2014 To Merza 2009)
- Mohammad MS, Hirotoshi I, Makiko N, Sun XS, Pham HN, Kiyofumi O. (2007). DnaJ gene sequence-based assay for species identification and phylogenetic prouping in the genus Staphylococcus. Int J. Sys Evol Microbiol, 57(1):25-30.
- Mohammed S H, Hmood M N, Abd A A, Obaid S A, Fahad B A, Kadhem F H. (2015). Screening of nasal carriage for Staphylococcus aureus and their resistance to oxacillin and cefoxitin among medical students in Karbala University. J Contemp Med Sci, 1(1), 13-6.
- Mumy KL and Findlay RH. (2004). Convenient determination of DNA extraction efficiency using an external DNA recovery standard and quantitative-competitive PCR. Journal of Microbiological Methods. Vol. 57, Issue 2, Pages 259- 268. https://doi.org/10.1016/j.mimet.2004.01.013
- Naorem, R. S., Urban, P., Goswami, G., & Fekete, C. (2020). Characterization of methicillin-resistant Staphylococcus aureus through genomics approach. 3 Biotech, 10, 1-19.
- Nusrat, J., Ifra, T. N., & Mrityunjoy, A. (2015). Detection of methicillin-resistant Staphylococcus aureus within raw milk and cheese samples. International Food Research Journal, 22(6), 2629.
- Oladipo I C, Ogunsona S B, Abayomi M A. (2019). Methicillin-resistant Staphylococcus aureus (MRSA) as a cause of nosocomial infection in Ibadan, Nigeria. European Journal of Pharmaceutical and Medical Research, 6(4), 135-139.
- Oladipo IC and Adejumobi OD. (2010). Incidence of Antibiotic Resistance in Some Bacterial Pathogens from Street Vended Food in Ogbomoso, Nigeria. Pakistan Journal of Nutrition; 9 (11): 1061- 1068.
- Oliveira D, Borges A, Simoes M. (2018). Staphylococcus aureus Toxins and Their Molecular Activity in Infections Diseases. Toxins. 10(6).
- Olowe, O., Eniola, K., Olowe, R. and Olayemi, A. (2007). Antimicrobial susceptibility and beta- lactamase detection of MRSA in Osogbo. South West of Nigeria. Nature and Science, 5, 44-48.
- Onemu OS, Ophori EA (2013). Prevalence of multi-drug resistant Staphylococcus aureus in clinical specimens obtained from patients attending the university of Benin teaching Hospital, Benin City, Nigeria. Journal of National Science Research, 3,154-9.
- Othman HE, Merza NS, Jubrael JMS. (2014). Nucleotide sequence analysis of methicillin resistant Staphylococcus aureus in kurdistan region-Iraq. Sci. J.Univ.Zakho. 2(1), 65-73.
- Rasigade JP, Vandenesch F. (2014). Staphylococcus aureus: a pathogen with still unresolved issues. Infection, genetics and evolution: journal of molecular epidemiology and evolutionary genetics in infectious diseases, 21:510-4.
- Sabri I, Adwan K, Essawi TA, Farraj MA. (2013). Molecular characterization of methicillin- resistant Staphylococcus aureus isolates in three different Arab world countries. European journal of microbiology & immunology. 3(3): 183-7.
- Sheagren JN. (1984). Staphylococcus aureus. The persistent pathogen. New Engl J Med; 310: 1368-73, 1437.
- Siegel, J. D., Rhinehart, E., Jackson, M., & Chiarello, L. (2007). Management of multidrug-resistant organisms in health care settings, 2006. American journal of infection control, 35(10), S165-S193.
- Silva V, Canica M, Capelo JL, Igrejas G, Poeta P. (2020). Diversity and genetic lineages of environmental Staphylococci: A surface water overview. FEMS Microbiol. Ecol, 96, fiaa 191.
- Skov, R., Smyth, R., Larsen, A. R., Bolmstrom, A., Karlsson, A., Mills, K., ... & Kahlmeter, G. (2006). Phenotypic detection of methicillin resistance in Staphylococcus aureus by disk diffusion testing and Etest on Mueller-Hinton agar. Journal of clinical microbiology, 44(12), 4395-4399.
- Taylor TA, Unakal CG. (2020). Staphylococcus aureus. StatPearls. Treasure Island ( FL) : StatPearls Publishing LLC.
- Tong SY, Davis JS, Eichenberger E, Holland TL and Fowler Jr VG. (2015). Staphylococcus aureus infections: epidemiology, pathophysiology, clinical manifestations, and management. Clinical microbiology reviews, 28(3), 603-661.
- Turner NA, Sharma- Kuinkel BK, Maskarine CSA, Eichenberger EM, Shah PP, Carugati M, et al. (2019). Methicillin- resistant Staphylococcus aureus: an overview of basic and clinical research. Nature reviews Microbiology., 17(4): 203-18.
- Yahya MQ, Azba SH and Al-Hayali MI. (2021). Effect of antibiotic misuse on the emergence of microbial resistance among urologic patients. Iraqi Journal of Pharmacy, 18(1), 44-56.
- Zapun A, Contreras-Martel C and Vernet T. (2008). Penicillin-binding proteins and β-lactam resistance. FEMS Microbiol. Rev 32:361-85.
QUICK LINKS
- SUBMIT MANUSCRIPT
- RECOMMEND THE JOURNAL
-
SUBSCRIBE FOR ALERTS
RELATED JOURNALS
- Journal of Genomic Medicine and Pharmacogenomics (ISSN:2474-4670)
- Food and Nutrition-Current Research (ISSN:2638-1095)
- Journal of Veterinary and Marine Sciences (ISSN: 2689-7830)
- Journal of Agriculture and Forest Meteorology Research (ISSN:2642-0449)
- Journal of Womens Health and Safety Research (ISSN:2577-1388)
- Journal of Astronomy and Space Research
- Journal of Genetics and Cell Biology (ISSN:2639-3360)






