Mini-Review
Genetic Structure of Corona Virus
3876
Views & Citations2876
Likes & Shares
Coronavirus is a group of RNA viruses that cause disease in mammals and birds. In humans and birds, they cause infections in the respiratory tract that vary from mild to fatal. Mild illnesses in humans include some cases of colds (which are also caused by other viruses, mainly rhinoviruses), while more deadly types can cause SARS, MERS, and COVID-19. They cause diarrhea in cows and pigs, while they cause hepatitis and encephalomyelitis in mice. There is still no vaccine or antiviral drug to prevent or treat coronavirus infection.
Introduction: The first reports of coronavirus infection in animals occurred in the late 1920s, when acute respiratory infections of domestic chickens appeared in North America. Arthur Schalke et al. Hawn gave the first complete report in 1931, identifying respiratory infections and lethargy with mortality rates above 40-90%.
Material and method: Inside the envelope is a nucleocapsid consisting of multiple copies of the nucleocapsid (N) protein, which bind to a single-stranded RNA single-stranded RNA genome in a continuous strand-like bead bond. Protein N is a 43 to 50 kDa phosphoprotein and is divided into three protected domains. Most proteins arecomposed of zones 1 and 2, which are typically rich in arginine and lysine. Domain 3 has a short carboxy terminal end and has a net negative charge due to excess acid over the amino acid residue. deducation: Coronavirus is a group of RNA viruses that cause disease in mammals and birds. The first reports of coronavirus infection in animals occurred in the late 1920s. Domain 3 has a short carboxy terminal end and has a net negative chargedue to excess acid over the amino acidresidue.
Keywords: RNA, SARS, Respiratory, Protein and carboxy
INTRODUCTION
The first reports of coronavirus infection in animals occurred in the late 1920s, when acute respiratory infections of domestic chickens appeared in North America [1-3]. Arthur Schalke and et al gave the first complete report in 1931, identifying respiratory infections and lethargy with mortality rates above 40-90% [4].
Leland David Bushnell and Carl Alfred Brandley isolated the infectious virus in 1933[5]. The virus was identified as infectious bronchitis virus (IBV). Charles Hudson and Fred Robert Budet first cultivated the virus in 1937 [6]. This sample was introduced as the Beaudette strain. In the late 1940s, two other coronaviruses, JHM, which causes brain disease (mouse encephalitis), and mouse hepatitis virus (MHV), which cause hepatitisin mice. It was not known at the time whether the three different viruses were linked [7, 8].
Human coronaviruses were discovered in the 1960s [9] using two different methods in the United Kingdom and the United States [10]. EC Kendall, Malcolm Bynoe, and David Tyrrell [11] who worked in the Cold Unit of the British Medical Research Council, nameda 1968 cold-specific virus called B814. Viruses cannot be successfully cultured using standard techniques for culturing viral viruses, adenoviruses, and so on. Known cold viruses were successfully transplanted in1965 by Tyrell and Bynoe through organ culture from the human embryonic trachea [12]. The new culture method was announced to the laboratory by Bertil Horn [13]. The isolated virus, when injected into a volunteer through the nose, causes them to catch a cold and is inactivated by ether, which indicates that it contains a fat pack. Dorothy Hamra. And John Proseno at the University of Chicago in 1962 separated a new cold from medical students. They isolated the virus in kidney tissue culture and
identified it as 229E. The new virus caused colds in the volunteers and was similarly inactivated by ether B814 [14].
Scottish virologist June Almeida at St Thomas' Hospital in London, in collaboration with Tyrrell, compared the structures of IBV, B814 and 229E in 1967[15] and showed that, using an electron microscope, the three viruses were as distinctin shape and cluster sas [16]. They are morphologically related to each other. A team at the National Institutes of Health in the same year was able to isolate another species of the new virus using organ culture and named one of the samples (OC43OC for organ culture) [17]. Like B814, 229E, and IBV, the new cold virus OC43 has distinct clusters, such as wood, when viewed under an electron microscope [18,19). It was soon shown that cold viruses, such as IBV, were similar in appearance to murine hepatitis virus. This group of new viruses was called coronaviruses after their distinctive appearance. Corona virus human virus 229E and coronavirus human virus OC43 continued to be studied in later decades [20]. Coronavirus B814 virus is gone [21]. It was not known if the human corona virus existed. Other human coronaviruses were identified, including SARS-CoV in 2003, HCoV NL63 in 2003, HCoV HKU1 in 2004, MERS-CoVin 2013, and SARS-CoV-2 in 2020 [22,23]. There have also been a large number of animal viruses. It has been identified since the 1960s [24].
MATERIAL & METHODS
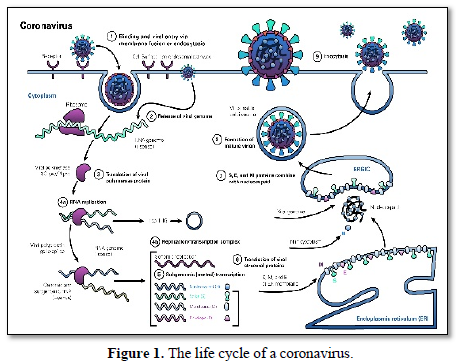
Replication cycle (Figure 1)
Viral infection begins when a viral spike protein binds to its receptor host cell receptor. After binding, the host cell protease is broken down and activates the receptor-bound spike protein. Depending on the existing host cell protease, degradation and activation allows the virus to enter the host cell by endocytosis or direct fusion of the viral coating with the host membrane.
Protein RdRp (Figure 2)
Distribution: RdRPs are very keeped throughout viruses and is even dependent to telomerase, though the reason for this is an ongoing inquiry as of 2009[25]. The similarity has led to speculation that viral RdRps are ancestral to human telomerase.
The most popular sample of RdRP is that of the poliovirus. The viral genome is composed of RNA, which enters the cell through receptor-mediated endocytosis. From there, the RNA is able to act as a template for suppplementary RNA synthesis, immediately. The supplementary strand is then, itself, able to act as a sample for the production of new viral genomes that are further packaged and released from the cell ready to infect more host cells. The advantage of this method of replication is that there is no DNA stage; replication is quick and easy. The disadvantage is that there is no 'back-up' DNAcopy.
Many RdRPs are affiliated tightly with membranes and are, so, hard to study. The best- known RdRPs are polio viral 3Dpol, vesicular stomatitis virus L and hepatitis C virus NS5B protein [26].
Many eukaryotes also have RdRPs involved in RNA interference; these furtify micro RNAs and small temporal RNAs and product double-stranded RNA using small interfering RNAs as primer [27] In fact these same RdRPs that are used in the defense mechanisms can be arrogate by RNA viruses for their benefit. Their evolutionary history has been reviewed [28].
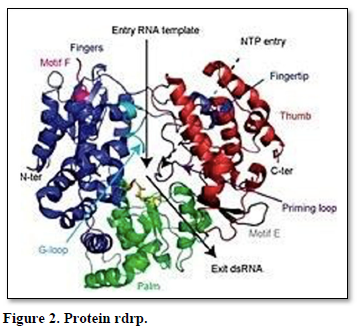

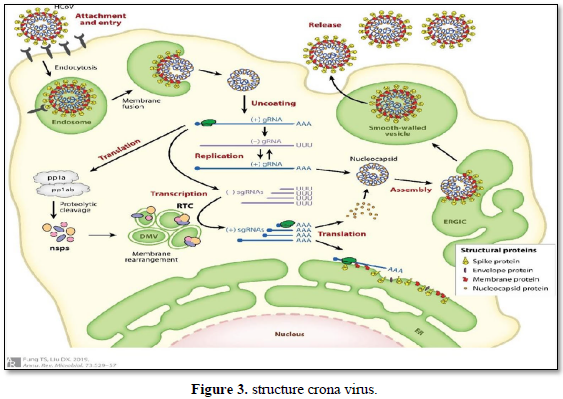
Structure (Figure 3): Viral/prokaryotic RNA-directed RNA polymerases, participant with many single- subunit DNA-directed polymerases, work a ply whose organization has been likened to the shape of a right hand with three subdomains termed fingers, palm, and thumb [29]. Only the palm subdomain, composed of a four-stranded antiparallel beta sheet with two alpha helices, is well keeped among all of these enzymes. In RdRP, the palm sub domain comprises three well-keeped motifs (A, B, and C). Motif A (D-x(4,5)-D) and motif C (GDD) are spatially juxtaposed; the aspartic acid residues of these motifs are implied in the binding of Mg2+ and/or Mn2+. The asparagine residue of motif B is involved in selection of ribonucleoside triphosphates over dNTPs [30].
The domain organization [31] and the 3D structure of the catalytic centre of a wide range of RdRPs, even those with a low overall sequence homology, are keeped. The catalytic centre is formed by many motifs containing a number of conserved amino acid residues.
Eukaryotic RNA interference requires a cellular RNA-dependent RNA polymerase (cRdRP) unlike the "hand" polymerases, they simulate simplified multi-subunit DNA- dependent RNA polymerases (DdRPs), specifically in the catalytic β/β' subunits, in that they use two sets of double-psi β-barrels in the active site. QDE1 in Neurosporacrassa, which forms a homodimer, is sample of such an enzyme [32].Bacteriophage homologues, such as yonO, appear to be closer to cRdRP than DdRP [26]. yonO is an RNA polymerase belonging to DNA [13].

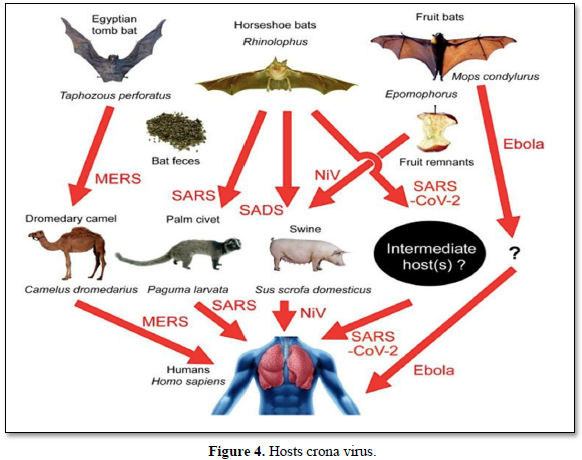
Hosts Corona virus (Figure 4)
Bat [33]: Researchers first began studying the virus genome to see if they could adapt to the pathogens found in other animals. In late January, a few weeks after researchers sequenced the SARS-CoV-2 genome, scientists at the Wuhan Institute of Virology Collected the entire corona virus sequence stored in their laboratory since the discovery in medium-sized horseshoe bats. Posted online [34]. In Yunnan Province in 2013. That genome, called RATG13, is 96% SARS-CoV-2 compliant, making it the closest known relative and indicating that the new virus originated in bats.
Pangolin [35]: Pangolin viruses were very far from the direct ancestors of SARS-CoV-2, but the fact that they are the only wild mammals known in addition to bats to date living with corona viruses such as SARS-CoV-2 Shows that they cannot be investigated as an intermediate source. Scientists are looking for coronaviruses in other animals as well. Aaron Irving, an infectious disease researcher at Duke-NUS Medical School in Singapore, says the ancestor of SARS-CoV-2 could lurk in tissue samples stored in the laboratory. "Many labs have samples in their freezers," hesays.
Cat [36]: FCOV is not the coronavirus that causes COVID-19 (which is called the acute coronary heart disease syndrome virus [SARS-CoV-2]).While the virus is transmitted from bats to humans, there is no evidence that FCoV is transmitted from cats to humans. Cats have receptors in their respiratory system to which SARS-CoV-2 binds. In the laboratory, cats can infect the original SARS virus and transmit it to other cats. They did not get sickand did not pass it on to the people. No cases of SARS-CoV-2 have been found in cats at present.
Dog [37]: In early March 2020, the World Health Organization announced that the coronavirus (COVID-19) was a global epidemic [38]. In the panic caused by the spread of the virus, people are concerned not only about their own health but also about the health of their dogs. According to the Centers for Disease Control, "there is no evidence that pets play a role in the spread of the coronavirus. Therefore, there is no justification for taking action against pets that may endanger theirwell-being."
Palmcivet [39]: Coronavirus 2019 (COVID-19) caused by Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2) is a public and international health emergency. Sofar, however, there are still differences of opinion about the source of the virus and its intermediate host. Here, we found that the corona virus was closely related to wildlife corona viruses including Pagumalarvata, Civet, and Rhinolophussinicus and was in the same phylogenetic tree branch. However, the genome and homology of ORF1a indicate that the virus is not the same as the corona virus that is the corona virus derived from these animals, while the virus has the highest similarity to the beta-corona virus isolated virusRaTG13.
Camel [40]: Coronavirus Respiratory Syndrome Virus was first found in the Middle East in late2012 in Saudi Arabia. Since then, there have been reports of human itarian cases, not only in Saudi Arabia but also in other countries in the Gulf region, such as the UAE, Qatar and Oman. To date, 2468 human cases of MERS-CoV have been reported from 27 countries worldwide (WHO, 2019) with a mortality rate of 34% (WHO, 2019). Camels are the main reservoir of animals MERS - CoV. Camel transmission to humans has been reported in many cases.

CONCLUSION
The first reports of coronavirus infection in animals occurred in the late 1920s, when acute respiratory infections of domestic chickens appeared in North America. Malcolm Baino and David Tyrrell, who worked in the Cold Unit of the British Medical Research Council, collected a special cold virus designated in 1968, designated B814 [41,42]. Viral/prokaryotic RNA-directed RNA polymerases, participant with many single-subunit DNA-directed polymerases, work apply whose organization has been likened to the shape of a right hand with three sub domains termed fingers, palm, and thumb.
However, the genome and homology of ORF1a indicate that the virus is not the same as the corona virus that is the corona virus derived from these animals, while the virus has the highest similarity to the beta-corona virus isolated virus RaTG13. Camels are the main reservoir of animals MERS - CoV. Camel transmission to humans has been reported in many cases.
Wikipedia (2020) Corona virus. Available online at: www.wikipedia.org/wiki/Coronavirus
-
Hagan WA, Bruner DW, Gillespie JH, Timoney JF, Scott FW, et al.(1988) Hagan and Bruner's microbiology and infectious diseases of domestic animals: With reference to etiology, epizootiology, pathogenesis, immunity, diagnosis, and antimicrobial susceptibility (1988), pp: 951.
-
Estola T (1970) Coronaviruses, a new group of animal RNA viruses. Avian Diseases 14(2): 330-336.
-
Fabricant J (1998) The early history of infectious bronchitis. Avian Diseases 42(4): 648-650.
-
Bushnell LD, Brandly CA (1933). Laryngotracheitis in chicks. Poultry Science 12(1): 55-60.
-
Decaro N (2011) Gamma coronavirus. In Tidona C, Darai G (eds.) Gamma coronavirus‡: Coronaviridae. The springer index of viruses. Springer pp. 403-413.
-
McIntosh K, Becker WB, Chanock RM (1967) Growth in suckling-mouse brain of "IBV-like" viruses from patients with upper respiratory tract disease. Proc Natl Acad Sci USA 58(6): 2268-2273.
-
Richmond C (2005) David Tyrrell. BMJ: British Medical J 330 (7505): 1451.
-
Kahn JS, McIntosh K (2005) History and recent advances in coronavirus discovery. Pediatr Infect Dis J 24(11 Suppl): S223-S227, discussion S226.
-
Monto AS (1984) Coronaviruses. Viral Infections of Humans Springer pp. 151-165.
-
Kendall EJ, Bynoe ML, Tyrrell DA (1962) Virus isolations from common colds occurring in a residential school. British Medical J 2(5297): 82-85.
-
Tyrrell DA, Bynoe ML (1965) Cultivation of a novel type of common-cold virus in organ cultures. British Medical J 1(5448): 1467-1470.
-
Tyrrell DA, Fielder M (2002) Cold wars: The fight against the common cold. Oxford University Press, pp: 93-95.
-
Forrest D, James K, Yuzenkova Y, Zenkin N (2017) Single-peptide DNA-dependent RNA polymerase homologous to multi-subunit RNA polymerase. Nat Commun 8: 15774.
-
Almeida J (2008) June Almeida (née Hart). BMJ 336(7659): 1511.
-
Almeida JD, Tyrrell DA (1967) The morphology of three previously uncharacterized human respiratory viruses that grow in organ culture. J Gen Virol 1(2): 175-178.
-
Arber W, Haas R, Henle W, Hofschneider PH, Jerne NK, et al. (1974) Current topics in microbiology and immunology/ergebnisse der mikrobiologie und immunitätsforschung. Current Topics in Microbiology and Immunology. Berlin, Heidelberg: Springer, pp: 8.
-
Mahase E (2020) COVID-19: First coronavirus was described in The BMJ in 1965. BMJ 369: m1547.
-
McIntosh K, Dees JH, Becker WB, Kapikian AZ, Chanock RM (1967) Recovery in tracheal organ cultures of novel viruses from patients with respiratory disease. Proc Natl Acad Sci USA 57(4): 933-940.
-
Myint SH (1995) Human coronavirus infections. The coronaviridae. The Viruses. Springer US, pp: 389-401.
-
Corman VM, Jores J, Meyer B, Younan M, Liljander A, et al. (2014) Antibodies against MERS coronavirus in dromedary camels, Kenya, 1992-2013. Emerg Infect Dis 20(8): 1319-1322.
-
Zhu N, Zhang D, Wang W, Li X, Yang B, et al. (2020) A Novel Coronavirus from Patients with Pneumonia in China, 2019. New England J Med 382(8): 727-733.
-
Fung TSF, Liu DX (2019) Human coronavirus: Host-pathogen interaction. Annu Rev Microbiol 73: 529-557.
-
de groot RJ, Baker SC, Baric R, Enjuanes L, Gorbalenya AE, et al. (2011) Family Coronaviridae. In King AM, Lefkowitz E, Adams MJ, Carstens EB, International Committee on Taxonomy of Viruses, International Union of Microbiological Societies. Virology Division (eds.) Ninth Report of the International Committee on Taxonomy of Viruses. Oxford: Elsevier, pp: 806-828.
-
Suttle CA (2005) Viruses in the sea. Nature 437(7057): 356-361.
-
Timm C, Gupta A, Yin J (2015) Robust kinetics of an RNA virus: Transcription rates are set by genome levels. Biotechnol Bioeng 112(8): 1655-1662.
-
Iyer LM, Koonin EV, Aravind L (2003) Evolutionary connection between the catalytic subunits of DNA-dependent RNA polymerases and eukaryotic RNA-dependent RNA polymerases and the origin of RNA polymerases. BMC Struct Biol 3: 1.
-
Zong J, Yao X, Yin J, Zhang D, Ma H (2009) Evolution of the RNA-dependent RNA polymerase (RdRP) genes: Duplications and possible losses before and after the divergence of major eukaryotic groups. Gene 447(1): 29-39.
-
Hansen JL, Long AM, Schultz SC (1997) Structure of the RNA-dependent RNA polymerase of poliovirus. Structure 5(8): 1109-1122.
-
Gohara DW, Crotty S, Arnold JJ, Yoder JD, Andino R, et al. (2000) Poliovirus RNA-dependent RNA polymerase (3Dpol): Structural, biochemical, and biological analysis of conserved structural motifs A and B. J Biol Chem 275(33): 25523-25532.
-
O'Reilly EK, Kao CC (1998) Analysis of RNA-dependent RNA polymerase structure and function as guided by known polymerase structures and computer predictions of secondary structure. Virology 252(2): 287-303.
-
Werner F, Grohmann D (2011) Evolution of multisubunit RNA polymerases in the three domains of life. Nat Rev Microbiol 9(2): 85-98.
-
Springer (2020) Available online at: www.link.springer.com
-
Hamre D, Procknow JJ (1966) A new virus isolated from the human respiratory tract. Proc Soc Exp Biol Med 121(1): 190-193.
-
Computational models, cell studies and animal experiments are being used to pinpoint the viral host that kicked off the pandemic (2020) Animal source of the coronavirus continues to elude scientists. Nature. Available online at: www.nature.com/articles/d41586-020-01449-8
-
Elwood vet (2020) Available onlie at: https://www.elwoodvet.net/coronavirus-in-cats
-
American Kennel Club (2020) Available online at: www.akc.org/expert-advice/news/can-dogs-get-coronavirus/
-
Knapp A (2020) The Secret History of the First Coronavirus. Forbes. Available online at: https://www.forbes.com/sites/alexknapp/2020/04/11/the-secret-history-of-the-first-coronavirus-229e/#4f2f5ec71d65
-
NCBI (2020) Available online at: www.ncbi.nlm.nih.gov
-
WHO (2020) Middle east respiratory syndrome. Available online at: www.who.int/news-room/fact-sheets/detail/middle-east-respiratory-syndrome-coronavirus- (mers-cov)
-
Lancet (2020) Available online at: www.thelancet.com
-
Vox (2020) Available online at: www.vox.com
QUICK LINKS
- SUBMIT MANUSCRIPT
- RECOMMEND THE JOURNAL
-
SUBSCRIBE FOR ALERTS
RELATED JOURNALS
- Journal of Blood Transfusions and Diseases (ISSN:2641-4023)
- Journal of Pathology and Toxicology Research
- Journal of Neurosurgery Imaging and Techniques (ISSN:2473-1943)
- International Journal of Internal Medicine and Geriatrics (ISSN: 2689-7687)
- International Journal of Medical and Clinical Imaging (ISSN:2573-1084)
- Journal of Ageing and Restorative Medicine (ISSN:2637-7403)
- Archive of Obstetrics Gynecology and Reproductive Medicine (ISSN:2640-2297)






