Review Article
Regulation of ULK2 Autophagy Activity by SGK1
3419
Views & Citations2419
Likes & Shares
CAPSULE
Background: UNC-21-like kinase 2 (ULK2) contains two putative phosphorylation residues (S507 and S750) by serum-and glucocorticoid-induced kinase 1 (SGK1).
Result: ULK2 is phosphorylated by SGK1.
Conclusion: ULK2 autophagy activity is negatively controlled by SGK1 phosphorylation.
Significance: ULK2 autophagy is up regulated during SGK1 inactivation, as an alternative cell survival pathway.
Serum- and glucocorticoid-induced kinase 1 (SGK1) is documented to have consensus sequence of phosphorylation site R-x-R-x-x-(S/T)-Ф, where Ф is any hydrophobic amino acid and arginine residues are conserved at positions -5 and -3 relative to positions of Ser/Thr residues that are phosphorylated in the presence of SGK1. With this information, it was noticed that UNC-21-like kinase 2 (ULK2) also harbors putative SGK1 phosphorylation sites at both S507 (502RsRnsSG508) and S750 (745RtRttSV751) residues. Thus, the purpose of this study was to discuss the biological significance of S507 and S750 residues of ULK2 phosphorylation sites by SGK1 as one of its authentic substrate proteins. Using ULK2 507 and 750 serine residue un- or phosphorylation analog (DA or DD), this modification of S507 or S750 residue seems to be required for the enhancement of ULK2 kinase activity. Thus, the phosphorylation at S750 or S507 residue seems to modulate its kinase activity, subcellular localization and protein interaction with AMPK1a, indicating that the phosphorylation of ULK2 by SGK1 regulates cell survival as an alternative modulation of its functions.
Keywords: ULK-2, SGK1, AMPK1a, mTORC1, Phosphorylation, Autophagy, Cell survival, Signal Transduction
INTRODUCTION
Similar with ULK1, uncoordinated 51-like kinase 2 (ULK2) is a member of the serine/threonine kinase protein family that plays an essential role in the regulation of autophagy in mammalian cells. ULK2 is expressed ubiquitously and involved in many fundamental biological processes, including cell fate determination, metabolism, transcriptional control, and autophagy (see the reviews 1-7). Even though its key role of autophagy in normal cellular homeostasis and multiple diseases has been studied, the specific functions of ULK1 and ULK2 in autophagy are currently unclear (8, 9). Because ULK2 is able to compensate for deletion of ULK1, its role seems to be redundant with that of ULK1. Even though ULK2 plays a central role in the autophagy signaling pathway, several recent studies suggest that the activity of ULK2 must be carefully regulated by mechanisms that are individually modulated for each substrate to avoid the indiscriminate phosphorylation [1-9].
While ULK1 is predominantly found in the cytosol under the confocal microscopy, ULK2 is mainly located in the nucleus, although it is also found in the cytosol [10]. The mechanism by which ULK2 is localized to the nucleus has been reported. Even though ULK2 does not have any recognizable short, basic, classic import, or export sequences, it contains PY-NLS sequences which contribute to its nuclear localization. This character is a distinguishable feature from that of ULK1. Because its localization is likely to be indirectly regulated through association with binding proteins, it was assumed that a binding protein can regulate the subcellular localization of ULK2 by inhibiting its nuclear export [10].
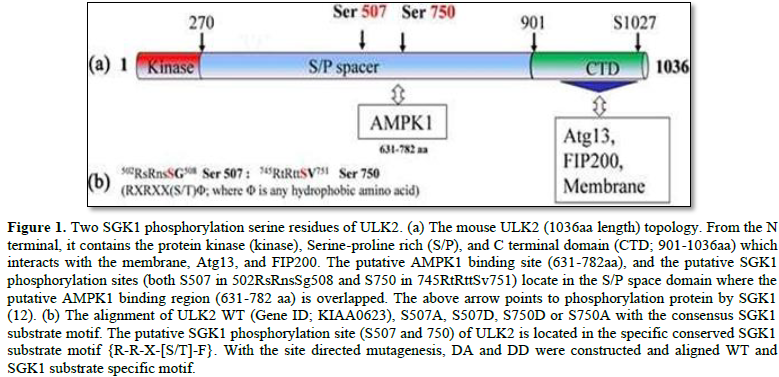
Both S507 (502RsRnsSG508) and S750 (745RtRttSV751) of ULK2 are noticed as the consensus Serum- and glucocorticoid-induced kinase 1 [SGK1] substrate sequences {R-x-R-x-x- [S/T] F} [11,12]. Thus, the objective of this review was to discuss whether SGK1 could phosphorylate S507 or S750 residue of ULK2 as one of its specific substrate proteins. Recently, it was documented that SGK1-mediated modification of S507 and/or S750 residue of ULK2 promoted its nuclear localization and autophagy activity with a synergic effect on its functional interactions with AMPK1a (13). Further, it was recognized that ULK2 expression was enhanced by treatment with GSK650394 (a SGK1 specific inhibitor) to compensate survival capacity by increasing its association with LC3 (13). In the case of activation of SGK1 by insulin treatment, ULK2 autophagy pathway was inhibited through phosphorylation of S507 or S750(12). Further, the researchers suggested that ULK2 autophagy seems to be an alternative cell survival pathway when PI3- PDK1-SGK1 pathway is inhibited (DA means S507A and S750A, while DD means S507D and S750D). Thus, in this review we discuss the role of the phosphorylation at S750 or S507 residue by SGK1 to modulate its kinase activity, subcellular localization and protein interaction with AMPK1a, as an alternative modulation of its functions [13].
The function roles of SGK1 phosphorylation on ULK2
Through determining whether S507 Ser and 750 residues change of ULK2 might be attributable to its subcellular localization or expression-level deviation, it was measured the Fraction ratio between nuclear and cytoplasm (Fn/c) value of each ULK2 construct under the confocal microscopy [10,13]. From their results, EGFP-ULK2 DD was the highest among three (ULK2 WT, DA, and DD), suggesting that the phosphorylation on ULK2 S507 and S750 residue by SGK1 promotes its nuclear localization (Figure 1).
To further elucidate the biological significance of ULK2 phosphorylation by SGK1, it was determined whether an endogenous ULK2 property change depending on SGK1 activator or inhibitor. Because LC3 binding motifs were also found in ULK2 S/P spacer (359tddfvlvphni360; Figure 2B), it was also determined whether endogenous ULK2 formed a protein complex with LC3 in HEK293 cells [3,5,6,10].
AMPK1a is known to be involved in the dual feedback (positive or negative) regulation of a variety of protein kinases [5,8,11]. AMPK1a is a conserved sensor of intracellular energy activated in response to low nutrient availability and environmental stress. Therefore, it was considered as a candidate protein that might interact with S/P space of ULK2 [10]. As shown in Figure 1, it has been reported that the S/P space domain of ULK2 WT protein interacted with AMPK1a [10,11]. To confirm this observation, it was performed coimmunoprecipitation of AMPK1a with EGFP ULK2 WT, S507A, DA, or DD in HEK293 cells. ULK2 DA mutant brought down AMPK1a than WT, S507A, and DD effectively (Figure 2), while ULK2 DD did less co-immunoprecipitated with AMPK1a in HEK 293 cells [13]. Thus, the result suggests that the phosphorylation on S507 and Ser705 residue of ULK2 inhibits the interaction between ULK2 and AMPK1a. The association with LC3II to ULK2 DA more strongly than WT or DD, suggested that ULK dephosphorylation at S507 and 750 residues could increase its binding to LC3II. Because LC3II binding indicates its autophagy activity, ULK2 DA mutant seems to contain more autophagy activity than ULK2 DD mutant or WT. The confocal microscopy observation was also performed with EGFP ULK2 WT, S507A, DA, or DD and AMPK1a antibody in HEK293 cells [13]. The high PCC value between ULK2 DA and AMPK1a was accounted, while PCC value between ULK2 DD and AMPK1a was low [11,13]. It seems to be that the interaction between ULK2 and AMPK1a is regulated by SGK1 phosphorylation [10,13].
Some intriguing features of the mechanism of ULK2 regulation by SGK1 phosphorylation on either S507 and/or S750 residues of ULK2 were indicated (Figures 1 and 2). In addition to a positive effect of the phosphorylation on both S507 and/or S750 of ULK2, a potential effect of autophagy with LC3II binding occurred via its action at an intracellular site in the C-terminus of the kinase mediated by adenosine monophosphate-activated kinase 1a [AMPK1a] binding (Figures 1 and 2) [8,11,13]. Thus, together with other reports, we indicated that the inhibition of ULK2 activity by S507 or S750 phosphorylation was agonized or antagonized by selective SGK1 activity modulators [13]. It was also reported that the basal activity of ULK2 S507A S750A (an analog of unphosphorylated ULK2) was higher than that of ULK2 WT or ULK2 DD (an analog of phosphorylated ULK2 by SGK1), suggesting that the S/P space domain of ULK2 near the S507 or S750 seems to be assigned in order to regulate its function by unknown mechanism beyond phosphorylation modification such as protein-protein interaction with AMPK1a or membrane [8,13].
With regard to ULK2, its interaction with AMPK1 is regulated competitively by SGK1- mediated phosphorylation of serine 507 and/or S750 residue within AMPK1 binding domain (Figure 2). ULK2 contains a consensus sequence for S750 phosphorylation by SGK1 within the AMPK1a binding domain. However, mutation of this serine residue, which is a prospective phosphorylation site within this domain in mutant ULK2 S750A, affected autophagy- dependent potentiation. Additionally, ULK2 (349tddfvlv356) association with LC3I is modified by phosphorylation on S750 [11]. Even though AMPK1a could interact with ULK2, other unknown proteins could also regulate ULK2 function through protein-protein interaction with its S/P space domain (Figure 1) [11,13]. Because AMPK1a is a conserved sensor of intracellular energy activated in response to low nutrient availability and environmental stress, the negative effect of SGK1 phosphorylation on the binding between ULK2 and AMPK1a suggested that SGK1 antagonizes AMPK1a for the regulation of ULK2 [8,11,13].
SGK1 has not been well characterized whether it has association with mammalian autophagy [12]. In this regard, SGK1 might essentially have the similar function as mTORC1 to inactivate ULK1/2 [12]. To control both cell autophagy and survival, these two kinases (mTORC1 and SGK1) appear to work together in a synergistic fashion. ULK2 S729 residue in (721kavlftvgSpphs733) which is close to ULK2 S750 is a putative phosphorylation site by mTORC1 [8,11]. Therefore, it is also worth noting that both SGK1 and mTORC1 are involved in the regulation of cell survival and autophagy synergistically [12]. Previously, it was documented that ULK2 contains PY-NLS which binds to karyopherin b2 for autophagy (783gaeaapslryvpy795) in its SP space domain, but not ULK1 [12]. Due to change of ULK2 conformation by SGK1 phosphorylation on its S750 residue (745RtRttSv751), the PY-NLS which is available to karyopherin b2 may eventually help transport it into the nuclear [11,13]. However, the specific function of ULK2 in the nuclear for autophagy is currently unclear. It still remains to be characterized whether ULK2 has its own specific biological role which is distinguishable to that of ULK1. Analysis of amino acid sequence did not reveal the presence of any recognizable import or export sequence. ULK2 subcellular localization may be indirectly regulated through association with binding proteins. ULK2 is located predominantly in the nuclei, although it is also present in the cytosol. However, the mechanism by which ULK2 localization is controlled remains unclear. Recently, it has been demonstrated that ULK2 is modulated through phosphorylation at its S1027 by Protein Kinase A (PKA), one of its authentic substrate proteins. As a consequence of PKA-phosphorylation on both S468 and S1027 of ULK2, its self-kinase activity on its 278-351 fragment and autophagy were reduced, coupled with an increase in sensitivity to starvation and stress, a promotion of its nuclear localization [12].
Because near the S1027 residue region, putative NES sequence form CRM1 binding site was noticed, the phosphorylation on S1027 of ULK2 by PKA may interfere its association with CRM1, and result in ULK2’s nuclear localization. ULK2 subcellular localization seems to be regulated by its specific serine/threonine residue phosphorylation by both SGK1 and PKA, similar with Figures 1 and 2. Several SGK1 substrates have been characterized and these results indicate that SGK1 plays a role, in concert with Akt1, in propagating effects of PI3K, including promotion of cell-cycle progression and consequently cell survival [11]. S507 and S750 residues of ULK2 also seems to be phosphorylated by Akt kinase which is a key protein kinase for cell survival [11-13]. Because ULK2 is involved in cell apoptosis, it seems to be reasonable that both SGK1 and Akt1 are antagonistic to ULK2 role by its S507 or S750 residue phosphorylation. ULK1 also contains a putative SGK1 phosphorylation site on S775 of (770RtRmfSA776) which is also characterized as AMPK1a phosphorylation site [11]. Therefore, SGK1 phosphorylation on S775 of ULK1 might also function, like AMPK1a (Figure 2). However, it is not reported yet whether the phosphorylation of ULK1 by SGK1 inhibits its autophagy activity. Future, it also required to be characterized that the phosphorylation on S507 or S750 of ULK2 by SGK1 (as its authentic substrate protein) is one of major regulatory points to activate its autophagy functions. Further, it has been reported that mTOR1 phosphorylation on S729 in (kavlftvgSpphs) motif of ULK2 also enhances ULK2’s autophagy activity, synergically [8,11].
To access further the effect of ULK2 phosphorylation by SGK1 on cell viability, it was measured the rate of apoptosis of HEK293 cell which was transfected with the EGFP ULK2 WT or its AA or DD mutant or EGFP vector through FACS [13]. and reported that EGFP ULK2 DA mutant showed significantly two times lower an apoptosis rate than ULK2 WT or ULK2 DD mutant construct. From these results, they suggested that the unphosphorylated ULK2 (DA mutant) contributes the cell survival effectively, while the phosphorylated ULK2 on its S507 and S750 residue by SGK1 (ULK2 DD) does not. The researchers also suggested that the association of ULK2 with LC3II is are regulated antagonistically, depending on the phosphorylation of serine residue of ULK2 by SGK1 [11,13]. Therefore, the phosphorylation on ULK2 S507and/or S750 residue by SGK1 also seems to control the cell survival (Figure 2).
ULK2 WT seems to be inhibited by SGK1 phosphorylation on its 507 and 750 serine residues. Through its phosphorylation, the functional proteins (such as Atg13-AMPK1a) are disassociated from ULK2 (left side). SGK1 activation through its upstream signal
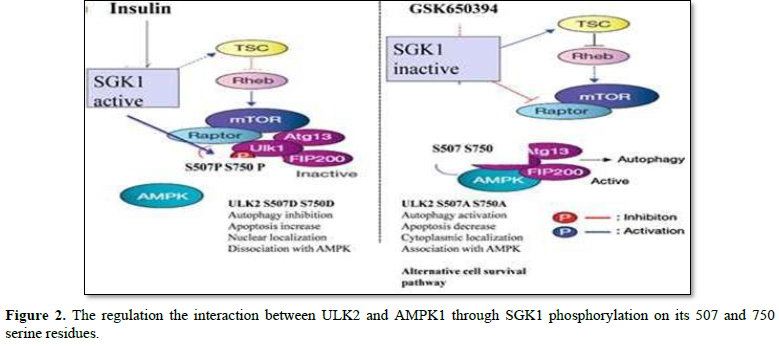


(such as insulin) blocks ULK2 autophagy activity through the promotion of AMPK1a dissociation, meanwhile SGK1 inhibition (by GSK650394) promotes ULK2 autophagy by the enhancement of AMPK1a association (8, 13). From the action mechanism of ULK1, this picture was redrawn by replacing it with ULK2. Even though ULK2 DD (which is an analogue of the phosphorylated ULK2) is localized into the nuclear and increase apoptosis, ULK2 AA (which is an analogue of the unphosphorylated ULK2 and is localized into the cytoplasm) is readily bound with AMPK1a, and active for the autophagy (more detail, see the discussion section).
FUTURE RESEARCH DIRECTIONS
Recent it was demonstrated that ULK2 is regulated through phosphorylation of its either S507 or S750 residue by SGK1 as one of its authentic substrate proteins. As a consequence of phosphorylation of S507 and/ or S750 residues of ULK2 by SGK1, its phosphorylation and association with AMPK1a were decreased [8,13], coupling with an increase of an antagonistic effect on its autophagy (Figure 2). SGK1 is also able to inhibit ULK2 autophagy activity by its phosphorylation as its authentic substrate proteins until they are active (Figure 2). Therefore, cell survival and autophagy are not antagonistic, but agonistic for cell survival in the transformed cell line. In the future, the signal pathways or protein kinases with ULK2 should be characterized to elucidate its delicate functions in the cell [8,10]. Moreover, the identification of each protein which interacts with ULK2 may also help to its unique role in the autophagy, distinguished with ULK1 (Figure 2).
ACKNOWLEDGEMENT
This work was supported by a Korean Research Foundation grant (BK21 plus and NRF- 2016R1D1A3B03934269) to S. S. Kang. Shin is Korean Research fellowship recipients (NRF- 2014R1A1A2009622).
- Velazquez AFC, Jackson WT (2018) So Many Roads: The Multifaceted Regulation of Autophagy Induction. Mol Cell Biol 38(21): e00303-e00318.
- Dodson M, Darley-Usmar V, Zhang J (2013) Cellular metabolic and autophagic pathways: traffic control by redox signaling. Free Radic Biol Med 63: 207-221.
- Mizushima N (2010) The role of the Atg1/ULK1 complex in autophagy regulation. Curr Opin Cell Biol 22(2):132-139.
- Noda NN, Mizushima N (2016) Atg101: Not Just an Accessory Subunit in the Autophagy-initiation Complex. Cell Struct Funct 41(1): 13-20.
- Wang B, Kundu M (2017) Canonical and noncanonical functions of ULK/Atg1. Curr Opin Cell Biol 45: 47-54.
- Alers S, Löffler AS, Wesselborg S, Stork B (2012) The incredible ULKs. Cell Commun Signal 10(1): 7.
- Chen Y, Liu XR, Yin YQ, Lee CJ, Wang FT, et al. (2014) Unravelling the multifaceted roles of Atg proteins to improve cancer therapy. Cell Prolif 47(2): 105-112.
- Fuqua JD, Mere CP, Kronemberger A, Blomme J, Bae D, et al. (2019) ULK2 is essential for degradation of ubiquitinated protein aggregates and homeostasis in skeletal muscle. FASEB J 33(11): 11735-11745.
- Chan EY, Kir S, Tooze SA (2007) siRNA screening of the kinome identifies ULK1 as a multidomain modulator of autophagy. J Biol Chem 282(35): 25464-25474.
- Shin SH, Lee EJ, Chun J, Hyun S, Kang SS (2015) ULK2 Ser 1027 Phosphorylation by PKA Regulates Its Nuclear Localization Occurring through Karyopherin Beta 2 Recognition of a PY-NLS Motif. PLoS One 10(6): e0127784.
- Yan J, Kuroyanagi H, Tomemori T, Okazaki N, Asato K, et al. (1999) Mouse ULK2, a novel member of the UNC-51-like protein kinases: Unique features of functional domains. Oncogene 18: 5850-5859.
- Kobayashi T, Deak M, Morrice N, Cohen P (1999) Characterization of the structure and regulation of two novel isoforms of serum- and glucocorticoid-induced protein kinase. Biochem J 344 Pt 1(Pt 1): 189-197.
- Shin SH, Lee EJ, Hyun S, Kang SS (2020) SGK1 Inhibits ULK2 Autophagy Activity. Am J Mol Biol 10: 12-28.




