Review Article
Glycan Heterogeneity on HIV-1 Env Decides the Fate of Virus Within Antigen Presentation Cells
5095
Views & Citations4095
Likes & Shares
HIV-1 envelope glycoprotein (Env) undergoes extensive post-translational glycosylation modifications cloaking the entire exposed Env surface with an assortment of high mannose, hybrid-, and complex-types of N-linked glycans (NLGs). This high density of NLGson HIV Env act as pathogen-associated molecular patterns (PAMPs) that enables the virion interaction with the host soluble and membrane bound carbohydrate-binding proteins (CBPs) particularly c-type lectin receptors (CLRs). CLRs with very fine and precise specificity to interact with the mannose rich glycans expressed on Env are expressed constitutively by various antigen-presenting cells (APCs) strategically located within the sexual route of virus transmission. Many of these CLRs like Langerin, DCIR, DC-SIGN, Mannose receptor (MR), BDCA2 and SIGLEC-1 play a very important role in the recognition, capture and dissemination of the virus within the host. The innate immune cells commonly use the CLRs to capture and internalize HIV for destruction and antigen presentation to T cells. However, HIV evades this CLR-mediated cellular degradation machinery and rather exploits it for transmission to CD4 T cells, the main cell types that host the robust and productive HIV replication. It is not known what determines the fate of virus whether being shuttled to degradative or trans-infection pathway of APCs. In this mini-review we address the relative importance of Env glycan composition and heterogeneity for HIV-1 transmission via the host membrane-associated carbohydrate-binding lectins particularly DC-SIGN and fate of virus being taken up by the cells expressing these CLRs.
Keywords: DC-SIGN, C-type lectin receptors (CLRs), Envelope glycoprotein, Viral degradation, Viral transmission, HIV-1 glycans
INTRODUCTION
Human Immunodeficiency Virus 1 (HIV-1) is the causative agent of Acquired Immunodeficiency Syndrome (AIDS). Despite continued and extensive research efforts, there is still neither a vaccine nor a cure for HIV/AIDS. This justifies a continuous search for novel and effective strategies for the effective treatment of this dreaded infection. The envelope glycoprotein (Env) like other enveloped viruses is the only molecular entity visible on the surface of HIV-1, embedded into the host derived lipid bilayer, mediating the first steps of cell attachment and entry through the primary receptor CD4 and co-receptor CCR5/CXCR4. The HIV-1 Env spike theoretically is the exclusive component of virus accessible to anti-HIV-1 neutralizing antibodies, thus, Env has been a target of intense research as a vaccine immunogen. Being central to viral pathogenicity and primary target of host immune response, the virus has developed extremely sophisticated mechanisms to shield Env spike glycoproteins, rendering it somewhat refractive to combating host defensive responses. HIV-1 Env is one of the known proteins with the highest genetic and antigenic variability. The distal most surface of HIV-1 Env is the main site of recognition by host receptors, which is decorated with poorly antigenic hypervariable loops (V1-V5). In addition, the Env glycoprotein (gp120/gp41 heterotrimer) is one of the most heavily glycosylated proteins found in nature, which is extensively covered with about 91 tightly packed potential N-linked glycosylation sites contributing approximately to 50% of the Env molecular mass [1-4]. The coating of the Env’s most exposed and antigenically vulnerable surface with poorly antigenic V1-V5 hypervariable loops, together with coating by extraordinary dense glycans which further shield the underlying antigenic surface, play a key role in the determination of Env structure and epitope exposure, which consequently affect the antigenicity, immunogenicity, antibody neutralization, infectivity and receptor binding [5-11]. This type of architecture of the Env with higher levels of variability and glycan shield is the strategy to escape the host selection pressure. However, this unusual extensive degree of HIV-1 Env glycosylation, taken as an immunologically silent shield, masking the preserved functional sites on the HIV-1for more than a decade, has emerged as an amazing target for recognition by broadly neutralizing antibodies (bnAbs) [12,13].
HIV INTERACTION WITH INNATE CELLS
Besides protection by formation of immunologically silent glycan shield, the HIV-1 has evolved to use these glycans to facilitate its infection of host. There is growing evidence that Env also plays a major role in the viral capture, transmission and dissemination during early stages of HIV-1 infection by hijacking the natural functions of the cells of innate immune system, the Langerhans Cells (LCs), Dendritic Cells (DCs) and macrophages, which are strategically located at all the entry sites like epidermis, mucosa, sub-mucosa, lymphoid and circulatory system to facilitate optimal interaction of the virus during sexual transmission, which is the most common cause of HIV infection [14,15]. The enigmatic features of these innate cell interactions with Env are mediated primarily by the membrane bound c-type lectin receptor (CLRs) family like Langerin, DC immunoreceptor (DCIR), DC-specific ICAM-3 grabbing non-integrin (DC-SIGN), Mannose receptor (MR), blood DC antigen 2 (BDCA-2) and Sialic acid-binding immunoglobulin-type lectins (SIGLEC-1), the most important CLRs which recognize exclusively the glycans on the surface of HIV-1 Env [16-22]. The innate cells and the CLRs they express, are spatially and temporally distributed along the sexual transmission pathway to generally form the first line of defense and perform differential functions leading finally to viral internalization and endosomal degradation for efficient antigen presentation, and modulate TLR-induced cytokine expression to enhance the infection of HIV.
C-TYPE LECTIN RECEPTORS
Carbohydrates are the natural ligands of CLRs. The CLRs recognize specific carbohydrate structures by means of one or more carbohydrate recognition domains (CRDs) and are grouped on the basis of the presence of a conserved structural motif in their CRDs. Various CLRs have distinguishable carbohydrate specificity, which are related to their amino acid sequence in their respective CRDs. The innate (DCs and macrophages) cell CLRs primarily interact with pathogens via the recognition of mannose, fucose and glycan carbohydrate structures. The HIV Env is known to be having highest glycan content and in particular has a very high proportion, ̴approximately 98% of glycans on native Env, as immature, unprocessed Man5-9GlcNAc2 type N-glycans, while the fully processed mature GlcNAc2Man3GlcNAc2 and the GlcNAcMan5GlcNAc2 containing N-glycans representing only upto 2% [5,19]. Together, this makes HIV-1 a susceptible target of these CLRs that recognize mostly high mannose type glycans [19,23,24]. Several of the CLRs encountered by HIV-1 in the sexual route like DC-SIGN, MR, Langerin and BDCA-2, exclusively recognize the high mannose glycans on gp120 of HIV-1, thus enhancing the recognition and capture of HIV-1 on these cells. These cells are potent antigen presenting cells that up-take the pathogens, process them and readily migrate to lymph nodes to present the antigen in conjunction with MHC molecules to the naïve CD4+ T-cells. These events are anticipated to play essential role in the initial events of HIV-1 transmission by transporting the virus from the peripheral mucosa to lymph node, the place with concentrated number of CD4+ T-cells, making the interaction of virus with the T-cells favorable with its ultimate targets [25-27].
INTERACTION OF HIV-1 WITH ANTIGEN PRESENTING CELLS AND SUBSEQUENT FATE OF VIRUS
There are two ways DCs direct the transmission of HIV-1 to CD4+T cells: DCs after capturing virus by CLRs, transfer captured virus in the absence of productive infection, which is referred to as in trans-infection or it endocytoses the virus within proteasome-resistant compartments in which the infectious virus is prevented from degradation and are released intact to infect the target cells (Figure 1) [28,29]. There are mounting evidences supporting the hypothesis that this hijacking of natural functions of DCs, degradation and presentation of antigens, by HIV-1 is determined by the glycan composition [30-33]. As mentioned, previously, the glycan composition on Env of different viruses is heterogeneous, as glycan maturation is primarily driven by the relative exposure of polypeptide, during folding, to Endoplasmic Reticulum (ER) and Golgi glycosylation modification enzymes. The specific constitution of Env between various HIV-1 strains may play a decisive role in a well-articulated CLR and DC-SIGN binding and transmission efficiency. A recent study showed the importance of the composition of HIV-1 glycans for DC-SIGN-mediated transmission [32-34]. The DC-SIGN as C-type (a type 2) lectin receptor (CLR) with a CRD, binds high-mannose glycans and fucose on the surface of viruses and bacteria, in a Ca2+-dependent manner. DC-SIGN displays enhanced affinity and specifically for high-mannose glycans with terminal Manα1-2 structures on HIV Env [25,35]. Although DC-SIGN serves as a PRR [15,36], yet the HIV-1 can evade the immune system mediated degradation machinery and exploit DC-SIGN for its transmission to CD4 T cells [15,30,34,37,38]. DC-SIGN interaction with HIV-1 is thus implicated in the initial stages of virus acquisition and spread from the mucosal site of virus entry [39]. However, the relative importance of these CLR expressing cells particularly DC-SIGN in HIV-1 transmission remains controversial. In some studies, antibody blockage of DC-SIGN on DCs was found to reduce virus capture only up to 50% [40,41], but other studies have demonstrated 70%-75% inhibition of virus capture by immature monocyte derived dendritic cells (MDDCs) and almost complete blockage of transmission to CD4 T cells [31]. The siRNA-based knock down of DC-SIGN in immature dendritic cells also been able to decrease the HIV-1 transfer to CD4 T cells by 75% [42].
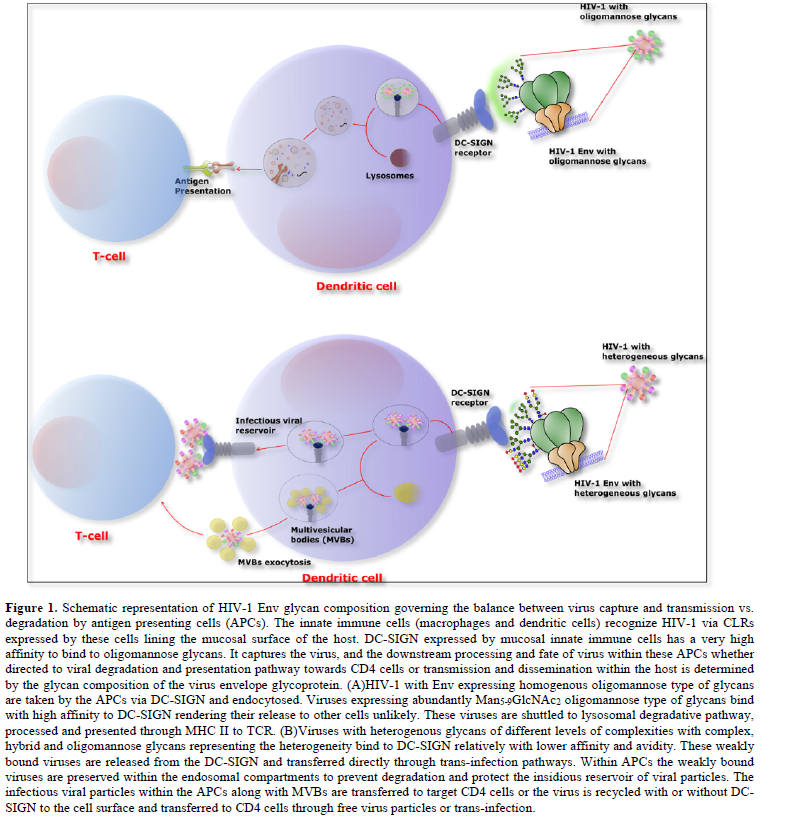

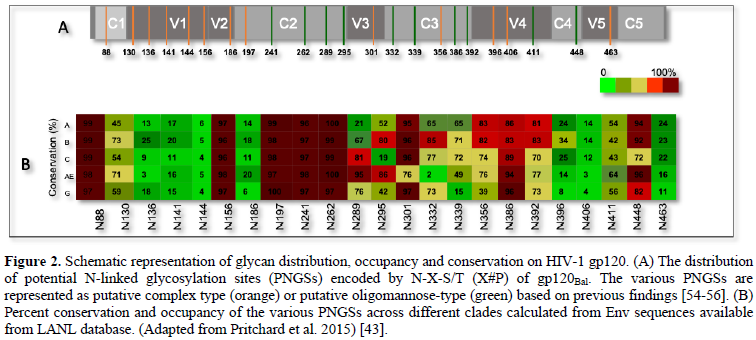
The glycan composition expressed on HIV Env is a key factor determining DC-SIGN-mediated virus transmission and fate of virus being taken up by the cells expressing these CLRs. Previously, it has been shown that HIV-1 enriched with high-mannose glycans was captured more efficiently by DC-SIGN and shuttled towards a degradation pathway, which augmented the MHCII-antigen presentation but impeded trans-infection [30]. The process of decorating surface of Env with glycans adds a layer of diversity to the already extremely high level of Env variation found among circulating HIV-1 isolates. At the genetic level, N-linked glycosylation is dictated by the N-X-S/T motif, where X is any amino acid except P. The Env proteins have varying number of these potential N-glycosylation sites (PNGSs), ranging from 23 to 34 per gp160 protomer; the majority of these are in the gp120 surface subunit, whereas only four to eight are in the external domain of the gp41 subunit [4]. Most PNGSs are not conserved. In fact, only six to eight PNGSs found in Env of clades A, B, C, G, and CRF_01.AE are >90% conserved, and for several PNGSs, conservation is <20% (Figure 2) [4,43]. The PNGSs on HIV-1 Env are not fully occupied [44,45]. The level of PNGS occupancy is dictated by site accessibility for a series of enzymes participating in the glycan maturation process [46]. The glycosylation pathway is initiated by the addition of Glc3Man9GlcNAc2en bloc onto a nascent protein in the ER. As the protein is transported across the ER and the Golgi apparatus, the high-mannose structure is trimmed and subsequently elaborated with hybrid- and complex-type glycans [19]. In contrast to cellular glycoproteins, which are usually adorned with mature complex-type glycans, virus Env carries all three glycan types, including early and intermediate high-mannose, intermediate hybrid, and mature complex glycans [47-49]. In fact, various glycan types and glycoforms are found in proportions that vary depending on Env strain and host cell type [4,23,44,45,50]. On soluble and membrane-anchored Env mimics, the high-mannose-type glycans range from 60–70%, with the most prominent being the least processed Man9GlcNAc2 glycans at 20–40% [19,47,48]. Analysis of soluble, uncleaved, prefusion-optimized BG505 Env gp140 trimers produced in 293F cells, similarly has shown that 56% are high-mannose type composed of Man5 (6%), Man6 (3%), Man7 (6%), Man8 (15%), and Man9 (26%) [51] hen the same proteins were produced in ExpiCHO cells, the total oligomannose content increased to 64%, with observable changes in glycoform proportions. Site-specific analysis further revealed that each PNGS on gp120 incorporated multiple glycoforms of only the oligomannose type or a mix of oligomannose and complex type, whereas the gp41 PNGSs had mainly complex type glycoforms [46,51].

The abundance of high-mannose glycans on HIV-1 Env and predominant binding of high-mannose glycans by DC-SIGN has been implicated in viral capture upon exposure and viral dissemination within the host. The interaction between HIV Env and DC-SIGN is influenced to a great extent by the glycan composition of virus [30,33,52]. Our recent findings also provide supporting evidence that virions carrying gp120 with higher numbers of oligomannose-type glycans are more efficiently endocytosed through DC-SIGN and more proficiently processed for antigen presentation than HIV-1 containing gp120 with heterogeneous glycans [34]. The transmission of oligomannose-enriched HIV-1 was relatively inefficient. Thus, the expression of oligomannose by HIV-1 enhances capture of DC-SIGN and transmission, but presence of heterogenous-glycansnegatively affect transmission by enhancing viral degradation (Figure 1). The mechanisms by which hybrid and complex glycans help HIV-1 evade degradation are not well understood yet. DC-SIGN has been shown to shuttle HIV-1 bearing high-mannose glycans to the degradative pathway, similar to other pathogens with such glycans [30]; this was indicated by an increased association with vesicles containing early and late endosomal markers and by more efficient antigen processing and presentation to MHCII-restricted CD4 T cells [30,34,53]. This study demonstrates that HIV-1 evades this degradative pathway by signals that prevent virus transport to endolysosomal compartments and preserve infectious virions on dendrites to promote their transfer to T cells [34].
- Lee JH, Ozorowski G, Ward AB (2016) Cryo-EM structure of a native, fully glycosylated, cleaved HIV-1 envelope trimer. Science 351: 1043-1048.
- Rudd PM, Dwek RA (1997) Glycosylation: Heterogeneity and the 3D structure of proteins. Crit Rev Biochem Mol Biol 32: 1-100.
- Jones GBS, Soto C, Lemmin T, Chuang GY, Druz A, et al. (2016) Trimeric HIV-1-Env Structures Define Glycan Shields from Clades A, B, and G. Cell 165: 813-826.
- Go EP, Ding H, Zhang S, Ringe RP, Nicely N, et al. (2017) Glycosylation benchmark profile for HIV-1 envelope glycoprotein production based on eleven env trimers. J Virol 91.
- Binley JM, Ban YE, Crooks ET, Eggink D, Osawa K, et al. (2010) Role of complex carbohydrates in human immunodeficiency virus type 1 infection and resistance to antibody neutralization. J Virol 84: 5637-5655.
- Crooks ET, Tong T, Chakrabarti B, Narayan K, Georgiev IS, et al. (2015) Vaccine-Elicited Tier 2 HIV-1 neutralizing antibodies bind to quaternary epitopes involving glycan-deficient patches proximal to the cd4 binding site. PLoS Pathog 11: e1004932.
- Kumar R, Tuen M, Liu J, Nadas A, Pan R, et al. (2013) Elicitation of broadly reactive antibodies against glycan-modulated neutralizing V3 epitopes of HIV-1 by immune complex vaccines. Vaccine 31: 5413-5421.
- Sanders RW, van Anken E, Nabatov AA, Liscaljet IM, Bontjer I, et al. (2008) The carbohydrate at asparagine 386 on HIV-1 gp120 is not essential for protein folding and function but is involved in immune evasion. Retrovirology. 5: 10.
- Townsley S, Mohamed Z, Guo W, McKenna J, Cleveland B, et al. (2016) Induction of heterologous tier 2 HIV-1-Neutralizing and cross-reactive v1/v2-specific antibodies in rabbits by prime-boost immunization. J Virol 90: 8644-8660.
- Wei X, Decker JM, Wang S, Hui H, Kappes JC, et al. (2003) Antibody neutralization and escape by HIV-1. Nature 422: 307-312.
- Reitter JN, Means RE, Desrosiers RC (1998) A role for carbohydrates in immune evasion in AIDS. Nat Med 4: 679-684.
- Lavine CL, Lao S, Montefiori DC, Haynes BF, Sodroski JG, et al. (2012) High-mannose glycan-dependent epitopes are frequently targeted in broad neutralizing antibody responses during human immunodeficiency virus type 1 infection. J Virol 86: 2153-2164.
- Walker LM, Huber M, Doores KJ, Falkowska E, Pejchal R, et al. (2011) Broad neutralization coverage of HIV by multiple highly potent antibodies. Nature 477: 466-470.
- Garcia-Vallejo JJ, van Kooyk Y (2013) The physiological role of DC-SIGN: A tale of mice and men. Trends Immunol 34: 482-486.
- Geijtenbeek TB, Torensma R, van Vliet SJ, van Duijnhoven GC, Adema GJ, et al. (2000) Identification of DC-SIGN, a novel dendritic cell-specific ICAM-3 receptor that supports primary immune responses. Cell 100: 575-585.
- .Useros NI, Lorizate M, Puertas MC, Plata MTR, Zangger N, et al. (2012) Siglec-1 is a novel dendritic cell receptor that mediates HIV-1 trans-infection through recognition of viral membrane gangliosides. PLoS Biol 10: e1001448.
- Lambert AA, Gilbert C, Richard M, Beaulieu AD, Tremblay MJ (2008) The C-type lectin surface receptor DCIR acts as a new attachment factor for HIV-1 in dendritic cells and contributes to trans- and cis-infection pathways. Blood 112: 1299-1307.
- .Zsolt DP, Perez JP, Erkizia I, Benet S, Pino M, et al. (2019) Dendritic Cells From the Cervical Mucosa Capture and Transfer HIV-1 via Siglec-1. Front Immunol 10: 825.
- Doores KJ, Bonomelli C, Harvey DJ, Vasiljevic S, Dwek RA, et al. (2010) Envelope glycans of immunodeficiency virions are almost entirely oligomannose antigens. Proc Natl Acad Sci USA 107: 13800-13805.
- Platt EJ, Wehrly K, Kuhmann SE, Chesebro B, Kabat D, et al. (1998) Effects of CCR5 and CD4 cell surface concentrations on infections by macrophagetropic isolates of human immunodeficiency virus type 1. J Virol 72: 2855-2864.
- Li Y, Svehla K, Mathy NL, Voss G, Mascola JR, et al. (2006) Characterization of antibody responses elicited by human immunodeficiency virus type 1 primary isolate trimeric and monomeric envelope glycoproteins in selected adjuvants. J Virol 80: 1414-1426.
- Li M, Gao F, Mascola JR, Stamatatos L, Polonis VR, et al. (2005) Human immunodeficiency virus type 1 env clones from acute and early subtype B infections for standardized assessments of vaccine-elicited neutralizing antibodies. J Virol 79: 10108-10125.
- Bonomelli C, Doores KJ, Dunlop DC, Thaney V, Dwek RA, et al. (2011) The glycan shield of HIV is predominantly oligomannose independently of production system or viral clade. PLoS One 6: e23521.
- Doores KJ (2015) The HIV glycan shield as a target for broadly neutralizing antibodies. FEBS J 282(24): 4679-4691.
- Feinberg H, Castelli R, Drickamer K, Seeberger PH, Weis WI, et al. (2007) Multiple modes of binding enhance the affinity of DC-SIGN for high mannose N-linked glycans found on viral glycoproteins. J Biol Chem 282: 4202-4209.
- van Liempt E, Bank CM, Mehta P, Vallejo JJG, Kawar ZS, et al. (2006) Specificity of DC-SIGN for mannose- and fucose-containing glycans. FEBS Lett 580: 6123-6131.
- Vallejo JJG, van Liempt E, da Costa Martins P, Beckers C, van het Hof B, et al. (2008) DC-SIGN mediates adhesion and rolling of dendritic cells on primary human umbilical vein endothelial cells through LewisY antigen expressed on ICAM-2. Mol Immunol 45: 2359-2369.
- Lekkerkerker AN, van Kooyk Y, Geijtenbeek TB (2006) Viral piracy: HIV-1 targets dendritic cells for transmission. Curr HIV Res 4: 169-176.
- Yokota YT, Muhsen M (2013) Development of human dendritic cells and their role in HIV infection: antiviral immunity versus HIV transmission. Front Microbiol 4: 178.
- van Montfort T, Eggink D, Boot M, Tuen M, Hioe CE, et al. (2011) HIV-1 N-glycan composition governs a balance between dendritic cell-mediated viral transmission and antigen presentation. J Immunol 187: 4676-4685.
- van Montfort T, Nabatov AA, Geijtenbeek TB, Pollakis G, Paxton WA, et al. (2007) Efficient capture of antibody neutralized HIV-1 by cells expressing DC-SIGN and transfer to CD4+ T lymphocytes. J Immunol 178: 3177-3185.
- Jan M, Upadhyay C, Pertejo JA, Hioe CE, Arora SK, et al. (2018) Heterogeneity in glycan composition on the surface of HIV-1 envelope determines virus sensitivity to lectins. PLoS One 13: e0194498.
- Jan M, Upadhyay C, Hioe CE (2019) HIV-1 envelope glycan composition as a key determinant of efficient virus transmission via DC-SIGN and resistance to inhibitory lectins. iScience 21: 413-427.
- Jan M, Arora SK (2017) Innate Sensing of HIV-1 by dendritic cell-specific ICAM-3 grabbing nonintegrin on dendritic cells: Degradation and presentation versus transmission of virus to t cells is determined by glycan composition of viral envelope. AIDS Res Hum Retroviruses 33: 765-767.
- Mitchell DA, Fadden AJ, Drickamer K (2001) A novel mechanism of carbohydrate recognition by the C-type lectins DC-SIGN and DC-SIGNR. Subunit organization and binding to multivalent ligands. J Biol Chem 276: 28939-28945.
- Geijtenbeek TB, Krooshoop DJ, Bleijs DA, van Vliet SJ, van Duijnhoven GC, et al. (2000) DC-SIGN-ICAM-2 interaction mediates dendritic cell trafficking. Nat Immunol 1: 353-357.
- Baribaud F, Doms RW, Pohlmann S (2002) The role of DC-SIGN and DC-SIGNR in HIV and Ebola virus infection: Can potential therapeutics block virus transmission and dissemination? Expert Opin Ther Targets 6: 423-431.
- Manel N, Hogstad B, Wang Y, Levy DE, Unutmaz D, et al. (2010) A cryptic sensor for HIV-1 activates antiviral innate immunity in dendritic cells. Nature 467: 214-217.
- Geijtenbeek TB, van Kooyk Y (2003) DC-SIGN: A novel HIV receptor on DCs that mediates HIV-1 transmission. Curr Top Microbiol Immunol 276: 31-54.
- Burleigh L, Lozach PY, Schiffer C, Staropoli I, Pezo V, et al. (2006) Infection of dendritic cells (DCs), not DC-SIGN-mediated internalization of human immunodeficiency virus, is required for long-term transfer of virus to T cells. J Virol 80: 2949-2957.
- Cambi A, de Lange F, van Maarseveen NM, Nijhuis M, Joosten B, et al. (2004) Microdomains of the C-type lectin DC-SIGN are portals for virus entry into dendritic cells. J Cell Biol 164: 145-155.
- Arrighi JF, Pion M, Garcia E, Escola JM, van Kooyk Y, et al. (2004) DC-SIGN-mediated infectious synapse formation enhances X4 HIV-1 transmission from dendritic cells to T cells. J Exp Med 200: 1279-1288.
- Pritchard LK, Spencer DI, Royle L, Bonomelli C, Seabright GE, et al. (2015) Glycan clustering stabilizes the mannose patch of HIV-1 and preserves vulnerability to broadly neutralizing antibodies. Nat Commun 6: 7479.
- Cao L, Pauthner M, Andrabi R, Rantalainen K, Berndsen Z, et al. (2018) Differential processing of HIV envelope glycans on the virus and soluble recombinant trimer. Nat Commun 9: 3693.
- Struwe WB, Chertova E, Allen JD, Seabright GE, Watanabe Y, et al. (2018) Site-Specific Glycosylation of Virion-Derived HIV-1 Env Is Mimicked by a Soluble Trimeric Immunogen. Cell Rep 24: 1958-1966 e5.
- Cao L, Diedrich JK, Kulp DW, Pauthner M, He L, et al. (2017) Global site-specific N-glycosylation analysis of HIV envelope glycoprotein. Nat Commun 8: 14954.
- Behrens AJ, Vasiljevic S, Pritchard LK, Harvey DJ, Andev RS, et al. (2016) Composition and Antigenic Effects of Individual Glycan Sites of a Trimeric HIV-1 Envelope Glycoprotein. Cell Rep 14: 2695-2706.
- Go EP, Herschhorn A, Gu C, Menendez LC, Zhang S, et al. (2015) Comparative analysis of the glycosylation profiles of membrane-anchored hiv-1 envelope glycoprotein trimers and soluble gp140. J Virol 89: 8245-8257.
- Kong L, Sheppard NC, .Jones GBES, Robson CL, Chen H, et al. (2010) Expression-system-dependent modulation of HIV-1 envelope glycoprotein antigenicity and immunogenicity. J Mol Biol 403: 131-147.
- Torrents de la Pena A, Rantalainen K, Cottrell CA, Allen JD, van Gils MJ, et al. (2019) Similarities and differences between native HIV-1 envelope glycoprotein trimers and stabilized soluble trimer mimetics. PLoS Pathog 15: e1007920.
- He L, Kumar S, Allen JD, Huang D, Lin X, et al. (2018) HIV-1 vaccine design through minimizing envelope metastability. Sci Adv 4: eaau6769.
- Guo Y, Feinberg H, Conroy E, Mitchell DA, Alvarez R, et al. (2004) Structural basis for distinct ligand-binding and targeting properties of the receptors DC-SIGN and DC-SIGNR. Nat Struct Mol Biol 11: 591-598.
- Gringhuis SI, den Dunnen J, Litjens M, van der Vlist M, Geijtenbeek TB, et al. (2009) Carbohydrate-specific signaling through the DC-SIGN signalosome tailors immunity to Mycobacterium tuberculosis, HIV-1 and Helicobacter pylori. Nat Immunol 10: 1081-1088.
- Cutalo JM, Deterding LJ, Tomer KB (2004) Characterization of glycopeptides from HIV-I(SF2) gp120 by liquid chromatography mass spectrometry. J Am Soc Mass Spectrom 15: 1545-1555.
- Go EP, Chang Q, Liao HX, Sutherland LL, Alam SM, et al. (2009) Glycosylation site-specific analysis of clade C HIV-1 envelope proteins. J Proteome Res 8: 4231-4242.
- Pabst M, Chang M, Stadlmann J, Altmann F (2012) Glycan profiles of the 27 N-glycosylation sites of the HIV envelope protein CN54gp140. Biol Chem 393: 719-730.
QUICK LINKS
- SUBMIT MANUSCRIPT
- RECOMMEND THE JOURNAL
-
SUBSCRIBE FOR ALERTS
RELATED JOURNALS
- Journal of Immunology Research and Therapy (ISSN:2472-727X)
- Journal of Clinical Trials and Research (ISSN:2637-7373)
- International Journal of Surgery and Invasive Procedures (ISSN:2640-0820)
- Oncology Clinics and Research (ISSN: 2643-055X)
- Journal of Cell Signaling & Damage-Associated Molecular Patterns
- Journal of Renal Transplantation Science (ISSN:2640-0847)
- International Journal of Clinical Case Studies and Reports (ISSN:2641-5771)




