6042
Views & Citations5042
Likes & Shares
Polytope construct sequence with linkers
HLWLTINEEAVIEAKCSNGNYTMEAVGNFTEMCSNGNYTMEAVGNFTECSFENGNYTMEAVGNFTCSHHLWLTINEEAVIEA
Determination of the attributes of the constructed polytope
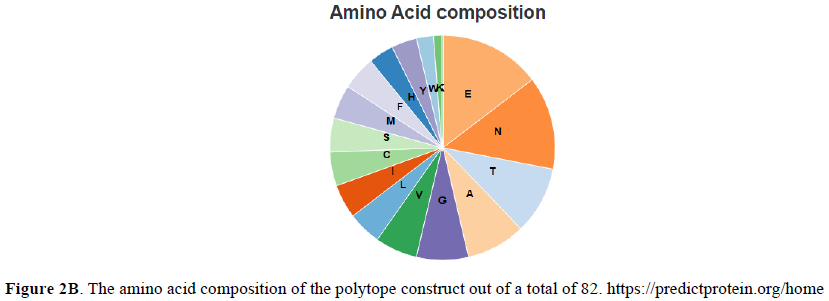
The propensity of the constructed polytope to be an antigen is determined by VaxiJenV2 online tool. The potential to be an allergen is evaluated by the online tool namely AllerTop V2. Similarly, the potential of the polytope to be toxic is assessed by ToxinPred online tool. Phyre2 and PredictProtein.org online tools are employed to construct its homology model (Figure 1) and secondary structure (Figure 2) respectively. The instability index, aliphatic index, GRAVY (Grand Average of Hydropathicity) and half-life of the polytope construct is evaluated using ProtParam tool Expasy (Table 2). PDBsum web server (http://www.ebi.ac.uk/thornton-srv/databases/cgi-bin/pdbsum/GetPage.pl?pdbcode=index.html) is used to generate a Ramachandran plot (Figure 3) for the constructed polytope to observe the position of amino acids in the Phi and Psi axis [13]. The PDBsum is a visual database that provides an overview of the residues of each 3D structure deposited in the Protein Data Bank format.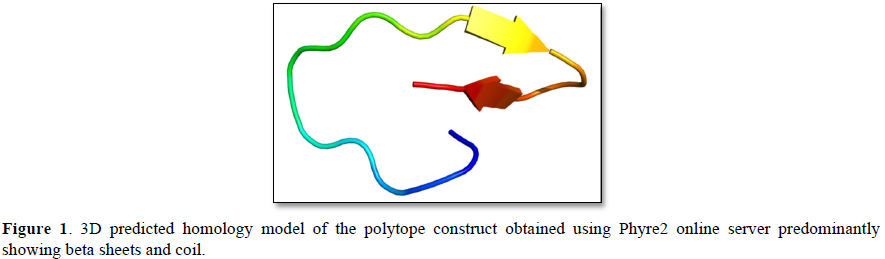

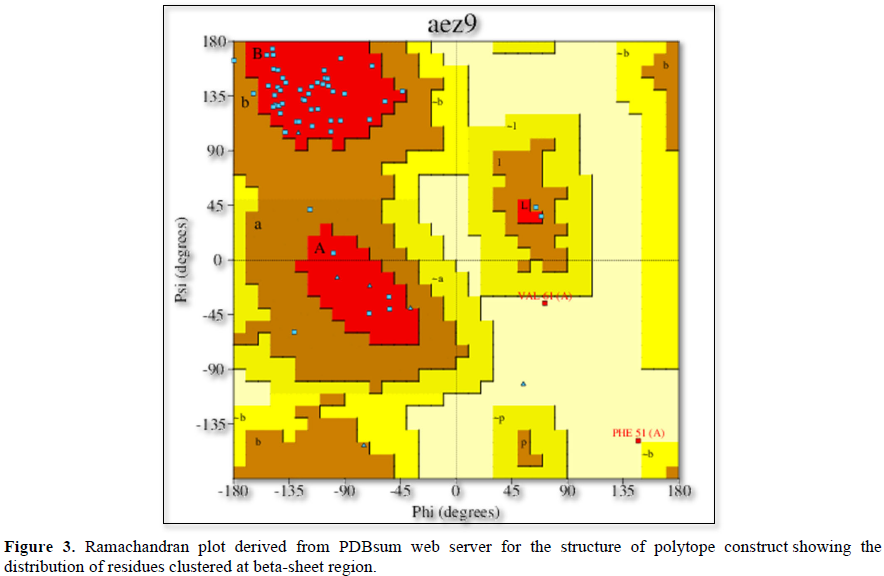
PROCHECK statistics (Table 3)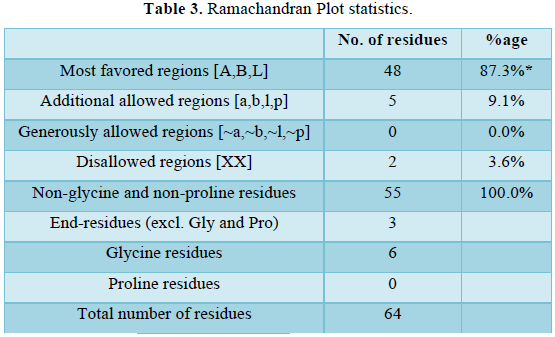
In-silico cloning of constructed polytope
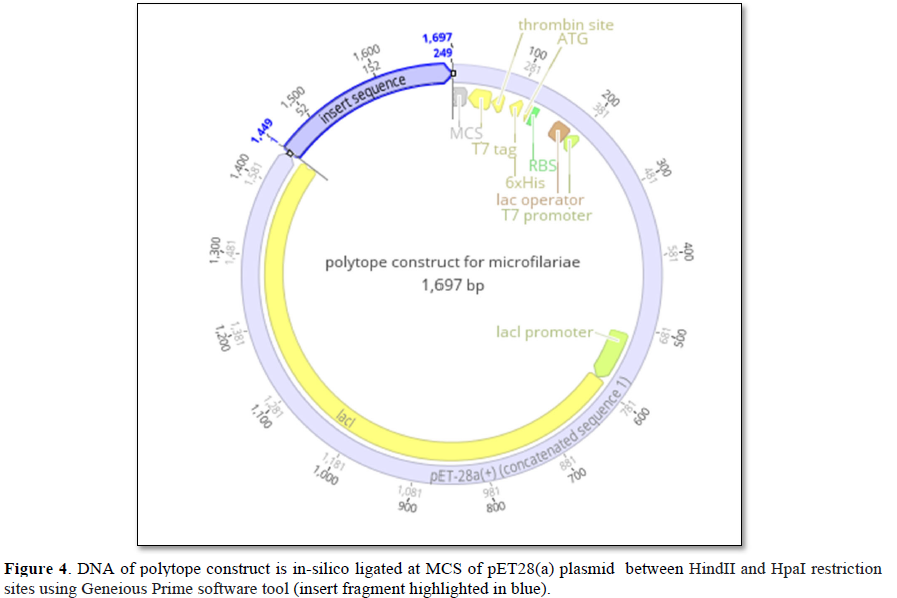
The amino acid sequence of the constructed Wb-Bhp-1 polytope is translated into corresponding DNA sequence using Expasy online server (https://web.expasy.org/translate/). The obtained 249 bp DNA sequence is in-silicol igated in between HindII and HpaI restriction sites at 3’ and 5’ directions in Geneious Prime online tool in MCS of pET28(a) plasmid which is selected as the expression vector with T7 promoter and kanamycin resistance (Figure 4). The constructed expression vector is to be transferred into the E.coli TOP10 cloning vehicle for in vitro generation of multiple copies of the polytope as a vaccine candidate.
Molecular Docking
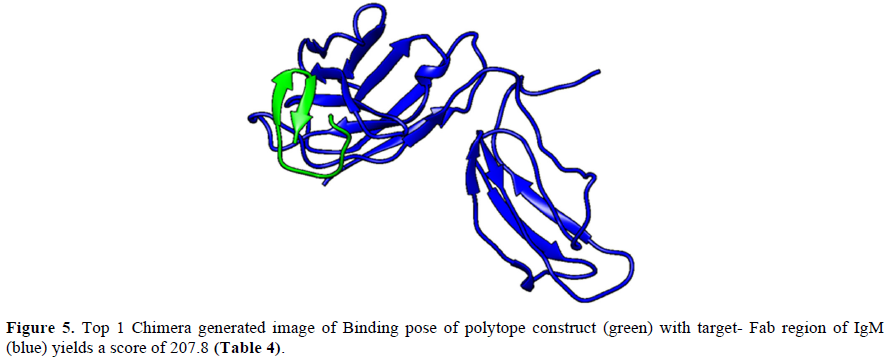
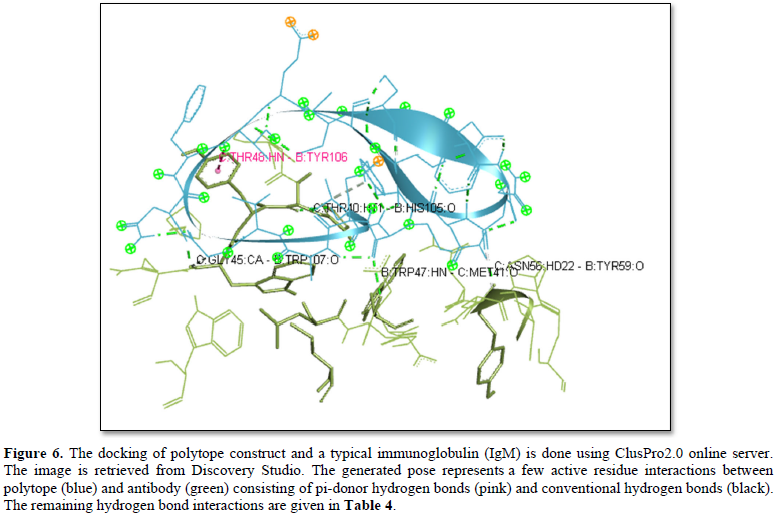
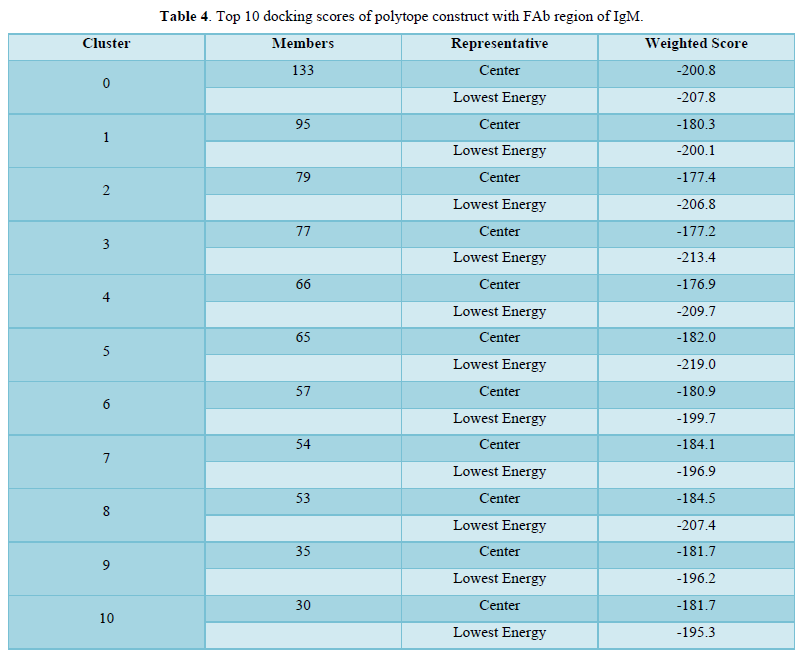
ClusPro 2.0 (https://cluspro.bu.edu/login.php?redir=/queue.php) web server is employed to perform docking of polytope construct against the Fab region of a typical IgM antibody (Figures 5 & 6). ClusPro 2.0 generates comparable results based on a large number of ligand/protein conformations; the scores of the top conformations interacting with the receptor grid are provided in ranked order (Table 4) [14,15].
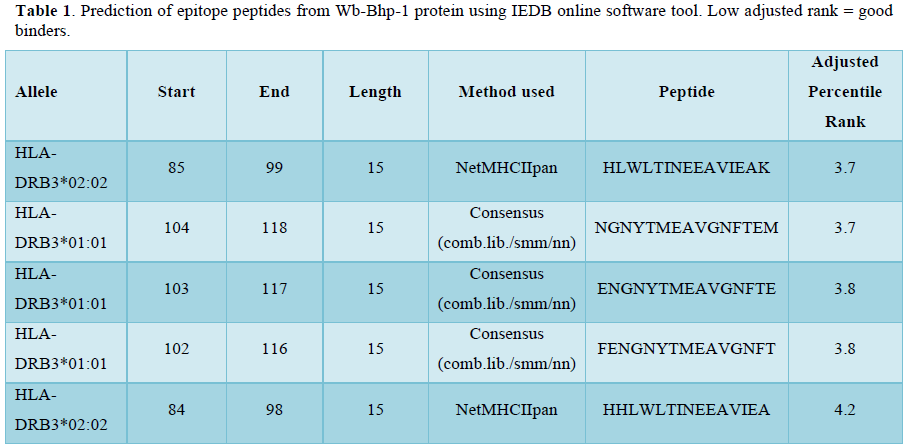
RESULTS AND DISCUSSION
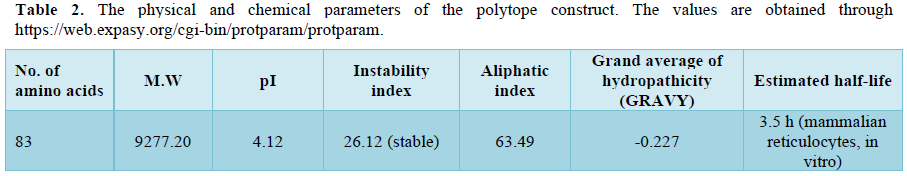
The amino acid sequence of the filarial antigen Wb-Bhp-1 developed by Greene et al [8] is used in the present study to derive the 15-mer epitopes in IEDB software tool which gave several numbers of combinatorial peptide sequences, of which, the Low adjusted rank peptides are reported to be good binders and such five are selected which are compatible to HLA class II DR B alleles and further constructed a polytope containing 82 amino acids with CS linkers (Table 1). The CS linkers are provided in the polytope to enable the same for proteolytic digestion in the proteasome of macrophage/dendritic cell by cysteine or serine proteases [2]. The physical and chemical attributes of the polytope construct reveal that the polytope is antigenic as authenticated by the VaxiJen V2 by yielding a value of 0.7456. Both AllerTop V2 and ToxinPred tools yielded the outcome as non-allergic and non-toxin respectively. Further, the polytope construct is found to be stable shown by an instability index value of 26.12, aliphatic index of 63.49 and an estimated half-life of 3.5 hours, sufficient period enough in the host to be identified by TLR2/4 [16] and to be encountered by a dendritic cell (Table 2).
The Phyre2 (Protein Homology/analogy Recognition Engine) and PredictProtein.org tools have yielded respectively the homology model (Figure 1) and the secondary structure of the constructed polytope. Upon submission in Phyre2 server revealed the presence of 4.0% alpha helix, 74% beta strands and 10% disordered which suggest that the polytope is a stable folded potential antigen to be chosen for the vaccine preparation. Furthermore, the PredictProtein.org yielded important observations on the quality of the polytope construct namely that the secondary structure is predominantly beta strand, exposed residues are significantly more compared to buried residues and protein binding residues are found to be predominant as shown in (Figure 2). The Ramachandran plot also revealed the distribution of residues in the most favored region clustered at beta-sheet region (Figure 3). The molecular docking of the polytope construct with the typical Fab of IgM revealed the binding pose with a score of 207.8 (Table 3) in ClusPro 2.0 indicating that the active residues (blue) have shown interaction with antibody (green) represented by pi-donor hydrogen bonds (pink) and conventional hydrogen bonds (black) along with the hydrogen bond interactions as given in Table 3 and Figures 5 & 6. The aforementioned attributes authenticate that the constructed polytope would be a vaccine candidate, which upon formulation will be used as a prophylactic regimen among the filarial endemic group.
To manufacture the vaccine candidate, the up-stream protocol requires a design of gene construct with a cloning vehicle. As shown in the Figure 4 the translated DNA of the polytope construct is in-silico ligated in pET28(a) plasmid which upon transferring into E.coli TOP10 cloning vehicle in a suitable bacterial growth medium will enrich the quantity of polytope which upon down-streaming steps, formulating with TLR2/4 agonist, stabilizers and preservatives, vaccine for filarial worm could be formulated for pre-clinical trials. The vaccine against filarial worm is quintessential requirement as elephantiasis is a disease with a serious social stigma in addition to illness. Further, microfilariae elicit IgG4 immune response [8,17,18]. The memory B-cells built will be a repertoire to neutralize Wb-Bhp-1 antigen of a freshly infected microfilariae which measures 177-230 μm and 5-7 μm in length and width respectively [19] and whose pre-patent period in bloodstream lasts several months [20] which would elicit both complement mediated and ADCC (antibody dependent cell cytotoxicity) response to arrest the intensity of lymphoedema.
ACKNOWLEDGEMENTS
The authors acknowledge VFSTR University for providing facilities through Centre of Excellence in the Department of Biotechnology.
- Roy N (2018) Elimination of lymphatic filariasis India Updates and way forward. Manipal J Med Sci 9: 1-3.
- Nutman TB (2001) Blood‐borne Filarial Infections: Wuchereriabancrofti, Brugiamalayi, Brugiatimori, Loa loa, Mansonellaperstans and Mansonellaozzardi. Principles and Practise of Clinical Parasitology pp: 433-455.
- Horton J, Witt C, Ottesen EA, Lazdins JK, Addiss DG, et al. (2000) An analysis of the safety of the single dose, two drug regimens used in programmes to eliminate lymphatic filariasis. Parasitology 121(S1): S147-S160.
- Babu S, Nutman TB (2014) Immunology of lymphatic filariasis. Parasite Immunol 36(8): 338-346.
- NVBDCP newsletter. Volume 4, Issue 4, April 2022. Available online at: https://nvbdcp.gov.in/Doc/Newsletter-13-May.pdf
- Cromwell EA, Schmidt CA, Kwong KT, Pigott DM, Mupfasoni D, et al. (2020) The global distribution of lymphatic filariasis, 2000-18: A geospatial analysis. Lancet Glob Health 8(9): e1186-e1194.
- Ottesen EA, Hooper PJ, Bradley M, Biswas G (2008) The Global Programme To Eliminate Lymphatic Filariasis: Health Impact After 8 Years. 2(10): e317.
- Greene SE, Fischer K, Choi YJ, Curtis KC, Budge PJ, et al. (2022) Characterization of a novel microfilarial antigen for diagnosis of Wuchereria bancrofti PLoS Negl Trop Dis 16(5): e0010407.
- Reynisson B, Alvarez B, Paul S, Peters B, Nielsen M (2020) NetMHCpan-4.1 and NetMHCIIpan-4.0: Improved predictions of MHC antigen presentation by concurrent motif deconvolution and integration of MS MHC eluted ligand data. Nucleic Acids Res 48(W1): W449-W454.
- Wang P, Sidney J, Dow C, Mothé B, Sette A, et al. (2008) A systematic assessment of MHC class II peptide binding predictions and evaluation of a consensus approach. PLoS Comput Biol 4(4): e1000048.
- Wang P, Sidney J, Kim Y, Sette A, Lund O, et al. (2010) Peptide binding predictions for HLA DR, DP and DQ molecules. BMC Bioinformatics 11: 568.
- Sreerama K (2021) Conserved envelope protein of nCoV2 as the possible target to design polytope vaccine. Explor Immunol 1: 155-165.
- Subramanian E (2001) Ramachandran GN. Nat Struct Mol Biol 8: 489-491.
- Vajda S, Yueh C, Beglov D, Bohnuud T, Mottarella SE, et al. (2017) New additions to the ClusPro server motivated by CAPRI. Proteins 85(3): 435-444.
- Kozakov D, Hall DR, Xia B, Porter KA, Padhorny D, et al. (2017) The ClusPro web server for protein-protein docking. Nature Protoc 12(2): 255-278.
- Fouzder C, Mukhuty A, Das S, Chattopadhyay D (2019) TLR Signaling on Protozoan and Helminthic Parasite Infection. In (Ed.), Toll-like Receptors. Intech Open.
- Prodjinotho UF, Hoerauf A, Adjobimey T (2019) IgG4 antibodies from patients with asymptomatic bancroftian filariasis inhibits the binding of IgG1 and IgG2 to C1q in a Fc-Fc-dependent mechanism. Parasitol Res 118(10): 2957-2968.
- Adjobimey T, Hoerauf A (2010) Induction of immunoglobulin G4 in human filariasis: An indicator of immunoregulation. Ann Trop Med Parasitol 104(6): 455-464.
- CDC (2018) Parasites - Lymphatic Filariasis. Biology - Life Cycle of Wuchereria bancrofti. Available online at: https://www.cdc.gov/parasites/lymphaticfilariasis/biology_w_bancrofti.html
- https://www3.paho.org/hq/index.php?option=com_content&view=article&id=5855:2011-general-information-lymphatic-filariasis&Itemid=4195&lang=en









