6968
Views & Citations5968
Likes & Shares
Similarly, RNA is chemically identical to DNA as it is a chain of similar monomers. RNA is further of three types: mRNA, tRNA and rRNA. RNA molecules are required at all stages of protein synthesis. Messenger RNA transmits the code that specifies the amino acid sequence of the protein; transfer RNA molecules translate the code word for word into protein; and ribosomal RNAs in the ribosome provide part of the machinery to perform the synthesis. Protein structure is known to have a higher degree of conservation compared to sequences due to large variations in sequence within the protein family which can still result in very similar three-dimensional structures [9]. The structure of any protein molecule helps in determining its function [10]. There are many important such findings and, we can analyze it by using different tools used in bioinformatics. Here we are discussing about some most common biological databases and tools used in sequence analysis of protein and nucleic acids.
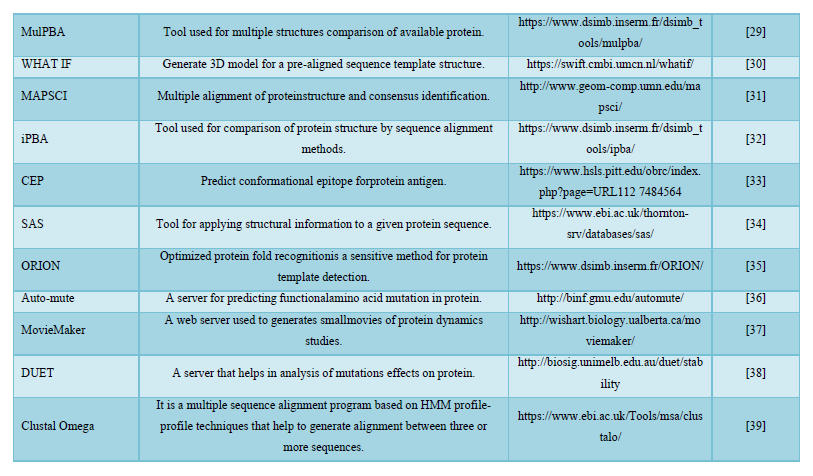
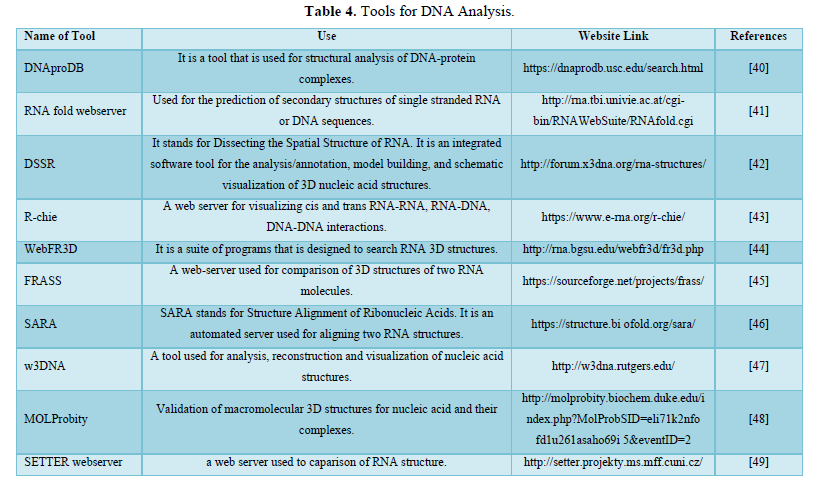
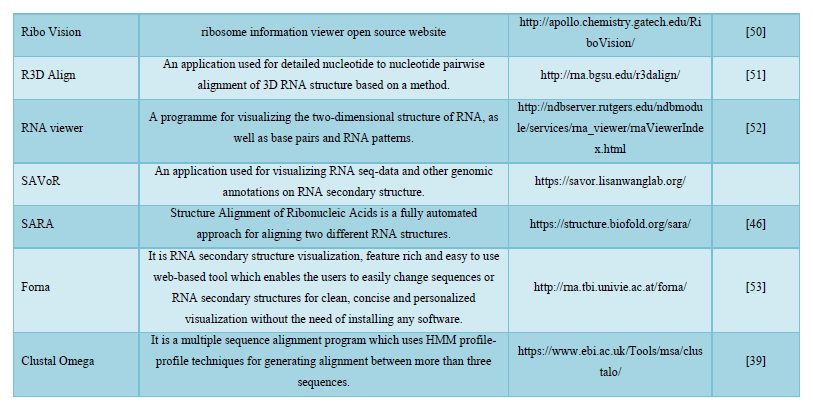
DISCUSSION
Bioinformatics is a discipline of biology that grows extensively in last few years. Sequences analysis and identification of new gene, proteins and structure are few important applications of bioinformatics [54]. The most important application is designing of 3D structure of proteins whose structures were not predicted by Nuclear magnetic resonance (NMR) and crystallographic method, due to protein bulky size and other limitations [55]. Genome analysis and sequencing of genome of new varieties is possible only because of extensive computational applications of bioinformatics. In this article we tried to compile most of the resources related to protein and nucleic acids that gives new insight to biological research [56-58].
CONCLUSION
Bioinformatics is a young discipline, which is widely used for analysis of genome, prediction of protein and gene structures, cell modeling, analysis of molecular pathways etc. As per the requirement of these tasks, various tools like the ones mentioned in this paper have been successfully curated and has made something as complex as genome sequencing much easier to work with. These tools can be used for various tasks like retrieval of structures, prediction and formation of new structures, comparison of different structures etc. that could be helpful for research of a new macromolecule. All these tools are easy to use and free to access.
- Benton D (1996) Bioinformatics - principles and potential of a new multidisciplinary tool. Trends Biotechnol 14(8): 261-272.
- Bayat A (2002) Science, medicine, and the future: Bioinformatics. BMJ 324(7344): 1018-1022.
- Bernstein FC, Koetzle TF, Williams GJ, Meyer EF, Jr Bric MD, et al. (1977) The Protein Data Bank. A computer-based archival file for macromolecular structures. Eur J Biochem 80(2): 319-324.
- Berman HM, Westbrook J, Feng Z, Gilliland G, Bhat TN, et al. (2000) The Protein Data Bank. Nucleic Acids Res 28(1): 235-242.
- Luscombe NM, Greenbaum D, Gerstein M (2001) What is bioinformatics? An introduction and overview. Yearb Med Inform 1: 83-99.
- Jorge NA, Ferreira CG, Passetti F (2012) Bioinformatics of Cancer ncRNA in High Throughput Sequencing: Present State and Challenges. Front Genet 3:
- Zagursky RJ, Russel D (2001) Bioinformatics: Use in Bacterial Vaccine Discovery. Biotechniques 31: 636-659.
- Minchin S, Lodge J (2019) Understanding biochemistry: Structure and function of nucleic acids. Essays Biochem 63(4): 433-456.
- Al-Karadaghi S (2022) Basic Principles of Protein Three-Dimensional Structure. Available online at: https://proteinstructures.com/structure/introduction/
- Alberts B, Johnson A, Lewis J, Raff M, Roberts K, et al. (2022) The Shape and Structure of Proteins. Available online at: https://www.ncbi.nlm.nih.gov/books/NBK26830/
- Nielsen M, Lundegaard C, Lund O, Petersen TN (2010) CPHmodels-3.0--remote homology modeling using structure-guided sequence profiles. Nucleic Acids Res 38: W576-W581.
- Lambert C, Léonard N, De Bolle X, Depiereux E (2002) ESyPred3D: Prediction of proteins 3D structures. Bioinformatics (Oxford, England) 18(9): 1250-1256.
- Kelley LA, Mezulis S, Yates CM, Wass MN, Sternberg MJ (2015) The Phyre2 web portal for protein modeling, prediction and analysis. Nat Protoc 10(6): 845-858.
- Guex N, Peitsch MC, Schwede T (2009) Automated comparative protein structure modeling with SWISS-MODEL and Swiss-PdbViewer: A historical perspective. Electrophoresis 30 Suppl 1: S162-S173.
- Roy A, Kucukural A, Zhang Y (2010) I-TASSER: A unified platform for automated protein structure and function prediction. Nat Protoc 5(4): 725-738.
- Kim DE, Chivian D, Baker D (2004) Protein structure prediction and analysis using the Robetta server. Nucleic Acids Res 32: W526-W531.
- Lamiable A, Thévenet P, Rey J, Vavrusa M, Derreumaux P, et al. (2016) PEP-FOLD3: Faster de novo structure prediction for linear peptides in solution and in complex. Nucleic Acids Res 44(W1): W449-W454.
- Chen CC, Hwang JK, Yang JM (2006) (PS)2: Protein structure prediction server. Nucleic Acids Res 34: W152-W157.
- Zemla A, Zhou CE, Slezak T, Kuczmarski T, Rama D, et al. (2005) AS2TS system for protein structure modeling and analysis. Nucleic Acids Res 33: W111-W115.
- Källberg M, Wang H, Wang S, Peng J, Wang Z, et al. (2012) Template-based protein structure modeling using the RaptorX web server. Nat Protoc 7(8): 1511-1522.
- Fiser A, Do RK, Sali A (2000) Modeling of loops in protein structures. Protein Sci 9(9): 1753-1773.
- Deprez, C, Lloubès R, Gavioli M, Marion D, Guerlesquin F, et al. (2005) Solution structure of the coli TolA C-terminal domain reveals conformational changes upon binding to the phage g3p N-terminal domain. J Mol Biol 346(4): 1047-1057.
- Berjanskii M, Tang P, Liang J, Cruz JA, Zhou J, et al. (2009) GeNMR: A web server for rapid NMR-based protein structure determination. Nucleic Acids Res 37: W670-W677.
- Ye Y, Godzik A (2003) Flexible structure alignment by chaining aligned fragment pairs allowing twists. Bioinformatics (Oxford, England) 19 Suppl 2: ii246-ii255.
- Maiti R, Van Domselaar GH, Zhang H, Wishart DS (2004) SuperPose: A simple server for sophisticated structural superposition. Nucleic Acids Res 32: W590-W594.
- Buchan DW, Minneci F, Nugent TC, Bryson K, Jones DT (2013) Scalable web services for the PSIPRED Protein Analysis Workbench. Nucleic Acids Res 41: W349-W357.
- Gelly JC, Brevern AG, Hazout S (2006) Protein peeling: An approach for splitting a 3D protein structure into compact fragments. Bioinformatics 22(2): 129-133.
- Negi S, Schein C, Oezguen N, Power T, Braun W (2007) InterProSurf: A web server for predicting interacting sites on protein surfaces. Bioinformatics 23(24): 3397-3399.
- Léonard S, Joseph A, Srinivasan N, Gelly J, de Brevern A (2013) mulPBA: An efficient multiple protein structure alignment method based on a structural alphabet. J Biomol Struct Dyn 32(4): 661-668.
- Vriend G (1990) WHAT IF: A molecular modeling and drug design program. J Mol Graph 8(1): 52-29.
- Ilinkin I, Ye J, Janardan R (2010) Multiple structure alignment and consensus identification for proteins. BMC Bioinformatics 11: 71.
- Gelly JC, Joseph AP, Srinivasan N, de Brevern AG (2011) iPBA: A tool for protein structure comparison using sequence alignment strategies. Nucleic Acids Res 39: W18-W23.
- Kulkarni-Kale U, Bhosle S, Kolaskar AS (2005) CEP: A conformational epitope prediction server. Nucleic Acids Res 33: W168-W171.
- Milburn D, Laskowski RA, Thornton JM (1998) Sequences annotated by structure: A tool to facilitate the use of structural information in sequence analysis. Protein Eng 11(10): 855-859.
- Ghouzam Y, Postic G, Guerin PEG, de Brevern AG, Gelly JC (2016) ORION: A web server for protein fold recognition and structure prediction using evolutionary hybrid profiles. Sci Rep 6(1): 28268.
- Masso M, Vaisman II (2010) AUTO-MUTE: Eeb-based tools for predicting stability changes in proteins due to single amino acid replacements. Protein Eng Des Sel 23(8): 683-687.
- Maiti R, van Domselaar GH, Wishart DS (2005) Movie Maker: A web server for rapid rendering of protein motions and interactions. Nucleic Acids Res 33: W358-W362.
- Pires DE, Ascher DB, Blundell TL (2014) DUET: A server for predicting effects of mutations on protein stability using an integrated computational approach. Nucleic Acids Res 42: W314-W319.
- Sievers F, Wilm A, Dineen D, Gibson TJ, Karplus K, et al. (2011) Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol Syst Biol 7(1): 539.
- Sagendorf JM, Berman HM, Rohs R (2017) DNA proDB: An interactive tool for structural analysis of DNA-protein complexes. Nucleic Acids Res 45: W89-W97.
- Gruber AR, Lorenz R, Bernhart SH, Neuböck R, Hofacker IL (2008) The Vienna RNA website. Nucleic Acids Res 36: W70-W74.
- Lu X, Bussemaker H, Olson W (2015) DSSR: An integrated software tool for dissecting the spatial structure of RNA. Nucleic Acids Res 43(21): e142.
- Lai D, Proctor JR, Zhu JY, Meyer IM (2012) R-CHIE: A web server and R package for visualizing RNA secondary structures. Nucleic Acids Res 40(12): e95.
- Sarver M, Zirbel CL, Stombaugh J, Mokdad A, Leontis NB (2008) FR3D: Finding local and composite recurrent structural motifs in RNA 3D structures. J Math Biol 56(1-2): 215-252.
- Kirillova S, Tosatto SC, Carugo O (2010) FRASS: The web-server for RNA structural comparison. BMC Bioinformatics 11: 327.
- Capriotti E, Marti-Renom MA (2009) SARA: A server for function annotation of RNA structures. Nucleic Acids Res 37(2): W260-W265.
- Lu XJ, Olson WK (2003) 3DNA: A software package for the analysis, rebuilding and visualization of three-dimensional nucleic acid structures. Nucleic Acids Res 31(17): 5108-5121.
- Chen VB, Arendall WB, 3rd Headd JJ, Keedy DA, Immormino RM, et al. (2010) Mol Probity: All-atom structure validation for macromolecular crystallography. Acta Crystallogr D Biol Crystallogr 66(Pt 1): 12-21.
- ČEch P, Svozil D, Hoksza D (2012) SETTER: Web server for RNA structure comparison. Nucleic Acids Res 40(W1): W42-W48.
- Petrov AS, Bernier CR, Hershkovits E, Xue Y, Waterbury CC, et al. (2013) Secondary structure and domain architecture of the 23S and 5S rRNAs. Nucleic Acids Res 41(15): 7522-7535.
- Rahrig RR, Petrov AI, Leontis NB, Zirbel CL (2013) R3D Align web server for global nucleotide to nucleotide alignments of RNA 3D structures. Nucleic Acids Res 41: W15-W21.
- Yang H, Jossinet F, Leontis N, Li C, Westbrook J, et al. (2003) Article navigation tools for the automatic identification and classification of RNA base pairs. Nucleic Acids Res 31(13): 3450-3460.
- Kerpedjiev P, Hammer S, Hofacker IL (2015) Forna (force-directed RNA): Simple and effective online RNA secondary structure diagrams. Bioinformatics (Oxford, England) 31(20): 337-3379.
- Robinson SW, Leader DP (2014) Bioinformatics: Concepts, Methods, and Data, in Handbook of Pharmacogenomics and Stratified Medicine.
- Fowler NJ, Sljoka A, Williamson MP (2020) A method for validating the accuracy of NMR protein structures. Nat Commun 11(1): 6321.
- Raux E, Schubert HL, Roper JM, Wilson KS, Warren MJ (1999) Vitamin b12: Insights into Biosynthesis’s Mount Improbable. Bioorg Chem 27(2): 100-118.
- Al-Karadaghi S, Hansson M, Nikonov S, Jönsson B, Hederstedt L (1997) Crystal structure of ferro chelatase: The terminal enzyme in heme biosynthesis. Structure 5(11): 1501-1510.
- Li F, Ryvkin P, Childress D, Valladares O, Gregory B, et al. (2012) SAVoR: A server for sequencing annotation and visualization of RNA structures. Nucleic Acids Res 40(W1): W59-W64.



