658
Views & Citations10
Likes & Shares
Nutrigenomics and
nutrigenetics is a new frontier in fish nutrition through dietary
supplementation with phytobiotics. A study was conducted to assess the effects
of supplementing Chromolaena odorata
leaves (COL) on the intestinal bacteria of Clarias
gariepinus fingerlings using 16S rRNA gene sequencing. One hundred and
eighty C. gariepinus fingerlings
(12.37 g, average weight) stocked in 12 net-happas (0.6 m × 1.1 m × 1.2 m)
suspended in an earthen pond (30 m × 5 m × 1.2 m) at 15 fish per net-happa
under four treatments in three replicates. Fish were fed four iso-nitrogeneous
diets containing: Control (0%), COL1 (0.5%), COL2 (1%) and COL3 (1.5%); ad libitum for 12 weeks. Molecular identification
of culture-dependent bacteria was carried out from the fish fed the COL in the
Department of Molecular Biology, Covenant University, Ota, Nigeria. Bacteria
counts of intestine were examined in using pour plate method on Nutrient and
MacConkey Agar. Bacterial isolates were subjected to DNA extraction,
amplification and purified PCR products were subjected to DNA gene sequencing.
Bacteria counts significantly differed (p<0.05) among the dietary
treatments. Total bacteria counts was highest in fish fed the control diet and
lowest in fish fed the 1% COL. Klebsiella
pneumoniae (COL1 and COL2), Proteus
mirabilis (COL3) and Shigella
flexneri (control) are the Enterobacteriaceae bacteria found in the fish
intestine based on the 16s rRNA gene sequencing. The study recommended the
dietary inclusion of C. odorata
leaves could effectively promote growth of cultured C. gariepinus by suppressing the growth of gram negative bacteria.
Keywords: Chromolaena
odorata, Bacteria, Clarias gariepinus,
Nutrigenetics, Gene sequencing
Abbreviations: DNA: Deoxyribonucleic Acid; RNA: Ribonucliec
Acid; PCR: Polymerase Chain Reaction; BLAST: Basic Local Alignment Search Tool;
dNTPs: di Nucleotides Proteins; FASTA: DNA and Protein Sequence Alignment
Software Package; MSA: Multiple Sequence Alignment; EMBL: European Molecular
Biology Laboratory; EBI: European Bioinformatics Institute; NCBI: National
Centre for Biotechnology Information
INTRODUCTION
Plants and weeds have been exploited for their medicinal value throughout
the world, Mukherjee and Wahile [1] reported the opinion of World Health
Organization, which stated that, ‘80% of the world’s population depend on
ancestral medicines for their haleness’. The authors further stated that: for
the health care of the remaining 20% population mainly residing in developed
countries, therapeutic product of plants and weeds play an important role.
Attempting to improve the quality of life, humans have used some of these
plants species as sources of food, shelter, clothing, and medicine, cosmetic
and for seeking relief from hardship of life. Thus, authors such as Sliva
Junior et al. [2] concluded that some of these plants are proven to be
medicinal because they contain some active substance that
Furthermore, Odunbaku and Ilusanya [4]
reported that searches for substance with antimicrobial activity and healing
activities are frequently considered interesting by some researchers since they
are frequently used in medicine as remedies for many infectious diseases. It
has also been reported by Hostettmann et al. [5] that the African continent has
a long history with the use of plants and in some African countries, close to
90% of the population relies on medicinal plants (phytobiotics) as a source of
drugs. Demand for phytobiotics, both local and international is continuously
growing, as well as the biological improving activities searching for sources
of new drugs [6]. Thus, these medicinal plants (phytobiotics) play a promising
role in aquaculture by enhancing the resistance of cultured fish against
diseases but their biological effects depend on various factors such as time,
dosage, method of administration and the physiological conditions of fish.
Chromolaena odorata (L) R.M. King and H. Robinson
(plate 2), common name: Siam weed a major perennial shrub which belongs to the
family Asteraceae and grows up to 7 m tall [7]. The active compounds present in
the leaves of C. odorata are in
phenolic compound, flavonoid, alkaloid and steroid. Akinmoladun et al. [8]
stated that C. odorata leaves could
be developed as natural antibacterial substance. These compounds have been
shown to disturb the function of cytoplasmic membrane and therefore have been
described as antibacterial substances [9,10]. Pelczar and Chan [11] reported
the activities of a phenolic compound present in C. odorata that at low concentration, phenolic compound damage
cytoplasm membrane causing the leakage of important metabolite and inactivated
bacterial enzymatic system. Also at high concentration, this compound can
damage cytoplasm cell membrane and to precipitate the cell protein and the
authors further stated that this compound interact with the component of
bacterial cell wall which causes higher permeability of the bacterial cell.
This compound also diffuses into the cell slowing and even stopping bacterial
growth. Moreover, Mori et al. [12] added that some flavonoid substances such as
robinetin, myricetin and epigallo catechin interfere with the intercalation
bond or hydrogen bond at the nucleic acid assembly preventing the activity of
DNA and RNA syntheses. Furthermore, steroid compound have been shown to prevent
microbial growth by damaging plasma membrane such that the cell cytoplasm was
leaked, thus causing cellular death [13].
Ghormade et al. [14] stated that the
emergence of nutrigenomics is to develop foods and feeds that could be linked
to genotypes of animals for a better production, productivity and health. Diet
on its own or by interaction with other environmental factors could cause
epigenetic changes that may turn certain genes on or off [15]. Harland [16] is
of opinion that the main role nutrients played in governing the cell content of
different proteins has been further investigated and a recognition of their
role as regulators of gene transcription, nuclear RNA processing, mRNA stability
and mRNA degradation (Ribonucleic Acid) has emerged. Nutrigenomics is a
prominent aspect due to its great potentiality for treating chronic disease, to
select animals for feed conversion efficiency, production and quality
improvement of products [15]. Garg et al. [17] stated that application of
genomic principles in a nutrigenomics assist in formulating a specific
association between nutrients and genetic factors. Also, nutrigenomics will
relate optimal diet to choose from many and different nutritional availability.
Nutrigenetics on the other hand, will provide information for identifying the
optimal diet for a given subject [18]. In order to define the optimal diets for
an individual’s; it is important to ascertain the health status at molecular stage
of the individual with the consideration of metabolic and epidemiological
studies. Nutri correlates diet, health and genomics in term of phenotypic
effect also include different -omics such as proteomics and transcriptomics
[19,20]. Different molecular and nutritional researches have shown that there
are several factors including environmental that are associated with animal
health.
To evaluating the interaction between diets
and genes, DNA microarray techniques and quantitative real-time Polymerase Chain
Reaction (PCR) and DNA sequencing are being employed [21]. These techniques
enable researchers to knowing the effects of nutrient which were impossible in
the past.
Molecular identification of bacteria culture
isolates is a modern technique used to determine the classification and
identification of bacteria using its deoxyribonucleic acid (DNA). This is
determining through gene sequencing analysis. It’s an approach in better
understanding and identification of bacteria. Molecular Identification of the work
of Ogawa et al. [22] investigated the facultative anaerobes by the 16S rRNA
sequence analysis. The basic local alignment search tool, BLAST program was
then used to determine homology with other organisms. Twenty-two strains were
isolated and identified as Enterococcus
faecalis or Enterococcus sp. from
the nutrient agar and GAM agar. Hence, this study entails the assessment of the
effect of C. odorata leaves on the
growth health status of C. gariepinus
through gene sequencing analysis of the intestinal bacteria of the fish.
MATERIALS AND
METHODS
Experimental site
The nutrition aspect of the study was carried
out at Aare Amoke Farms, Ifo, Ogun State, Nigeria, which is located within the
Southwestern part of Nigeria at 6°49΄ 00°N and 3°12΄ 00°E.
Experimental fish
A total of one hundred and eighty African mud
catfish (C. gariepinus) fingerlings,
purchased at a reputable farm was used as the test fish species in the study.
Preparation and
processing of C. odorata (whole
leaves preparation)
Fresh young leaves of C. odorata harvested within the premises of the Federal University
of Agriculture, Abeokuta, and Ogun State, Nigeria and were authenticated in the
Department of Forestry and Wildlife where a voucher specimen was maintained for
the plants: C. odorata ID No
UAHA/08.180002. The leaves were gotten by harvesting the whole plant above a 3 cm
stubble height within the mid-vegetative to early flowering stage of
development [23]. Thereafter, each leaf was hand-plucked from stems and placed
directly into jute-bag. The leaves C.
odorata were thoroughly rinsed with distilled water to remove dirt and
weighed on an electronic scale, cut into smaller pieces with the aid of a
knife. The leaves was blended with water which was added at a ratio of 1:1 (1 g
of the leaves is equivalent to 1 ml of water.) in a household electric blender
(Century, CB-8231-M, China), poured in a glass container and stored in the
refrigerator before adding into the basal diets [24].
Experimental system
The experiment was conducted at the Earthen Pond
Unit of dimensions 30 m × 5 m × 1.2 m (LBH) of Aare Amoke Farms, Ifo between
February 5th-May 20th, 2018. The study was conducted in
twelve (12) net-happa of dimensions 0.6 m × 1.1 m × 1.2 m (LBH) which were
suspended in the pond supplied with fresh water. One hundred and eighty
fingerling were randomly (completely randomized design) stocked to four (4)
treatments in the net-happa at a stocking rate fifteen fish per net-happa in
three replicates.
Four isonitrogenous diets were formulated at
40% crude protein to containing three varying levels of C. odorata leaves-paste and the control (Table 1). The samples of C.
odorata leaves paste with the three levels (0.5%, 1.0% and 1.5%) were added
on top of the basal diets and thoroughly mixed with the use of a mixer.
Compounded feeds were pelletized (2 mm) using
the pelletizing machine from the feed milling factory, dried to 10% moisture
content with the use of a dryer and allowed to cool in an open-air. Feeds were
later crushed into smaller particles based on the size of the fingerlings,
packed and stored in an opaque nylon bag according to the treatments.
Experimental procedure
The fish was weighed individually at the beginning
of the experiment and were acclimated to the experimental system for fourteen
days before the commencement of the study and fed two times daily with a
commercial diet (40% C.P.).
Fish feeding and
monitoring of growth
Before commencing the experiment, fish were
starved for 24 h to increase the appetite of the fish and were fed with the
diets at two feeding regimes, in the morning (08:00-09:00) and evening
(17:00-18:00), ad libitum for 12
weeks. Fish were weighed in each net-happa weekly using a sensitive electronic
weighing scale (Mettler Toledo FB602) for the growth of fish and ensuring that
the fish are consuming the feeds.
Determination of
intestinal bacteria of C. gariepinus
fingerlings
After the feeding study, fish were selected
from each of the treatment supplemented with C. odorata and the control for the molecular study. Dissection of
the midline in ventral surface of the fish was carried out to remove the gut
proximal section. 0.1 g of was sectioned from the fish intestine. This was
conducted at the Department of Microbiology, College of Science and Technology,
Covenant University, Ota, Ogun state.
The microbial counts and biochemical
identification of the bacteria were carried out as follows:
a. 1 g
of fish intestine was put into 1 ml of peptone water and incubated for 4 h
after which serial dilution was done up to the fifth dilution.
b. 1 ml
of each dilution was plated using pour plate method on Nutrient Agar and
MacConkey Agar.
c.
The plates were incubated for 24-48 h
after which the colonies were counted using colony counter (Model KA00-74A,
Vision Scientific, Japan). Colony forming unit were calculated for each count.
The samples were all done in duplicates.
d. The
colonies counted were put on slant for biochemical test. Biochemical test were
carried out according to Chessbrough [25] while the identification was done
using Bergey’s manual of Bacteriology and Manual for the identification of
medical Bacteria by Cowan and Steel [26].
Molecular
identification of bacteria isolated from the intestine of C. gariepinus fingerlings using 16S rRNA gene sequencing
The molecular identification of bacteria
isolated from fish intestine using 16s rRNA sequencing were carried out at the
Department of Molecular Biology, College of Science and Technology, Covenant University.
Five bacteria cultures isolated from the biochemical identification were put
forward for the molecular characterization. The stages involved in the
molecular characterization of the bacterial isolates are DNA extraction, 16S
rRNA Amplification (PCR analysis) and sequencing analysis.
DNA extraction from
bacterial cultures isolates
Bacteria possess plasma membrane, a rigid
cell wall and an outer membrane. All these mechanical barriers have to be
disrupted to release cellular component mainly DNA [27]. Ethylene
diaminetetraacetic acid (EDTA) chelates the bivalent ions present in the lipid
bilayer weakening the membrane. Sodium Dodecyl Sulphate (SDS) is used to
disrupt all membranes followed by the treatment of Chloroform: Isoamylalcohol
to denature protein and also to separate SDS and organic phases. Proteinase K
is added to denature protein in cell membrane. RNase A is added to avoid RNA
contamination. Chilled Isopropanol treatment causes precipitation of DNA from
aqueous phase. Precipitation with 70% ethanol eliminates divalent cations. DNA
obtained is suspended in TE buffer [28]. DNA from bacterial growth was
extracted using the DNeasy Blood and Tissue Kit (QIAGEN; Bechman Instruments
Inc., Triton). The following step by step procedures and protocols were
observed:
I.
1.5 mL bacteria culture cells was
transferred in 2 mL eppendorf tube aseptically and centrifuged for 5 min at
10,000 rpm. Supernatant was drained and the pellet was resuspended in 200 μl
PBS (pH 7.2, 50 mM potassium phosphate, 150 mM NaCl).
II.
20 μl proteinase K was added and 200 μl
Buffer AL was also added. The tube was mixed thoroughly by vortexing and
incubated at 56°C for 10 min.
III.
200 μl ethanol (96-100%) was added to
the sample, and mix thoroughly by vortexing again. The resulting mixture was
pipetted into the DNeasy Mini spin column placed in a 2 ml collection tube,
centrifuged at 8000 rpm for 1 min. The flow-through and collection tube was
discarded.
IV.
The DNeasy Mini spin column was placed
in a new 2 ml collection tube and 500 μl Buffer AW1 was added and centrifuge
for 1 min at 8000 rpm. The flow-through and collection tube was discarded.
V.
The DNeasy Mini spin column was placed
in a new 2 ml collection tube, and 500 μl Buffer AW2 was added and centrifuge
for 3 min 14,000 rpm to dry the DNeasy membrane. Flow-through and collection
tube was also discarded.
VI.
The DNeasy Mini spin column was placed
in a clean 1.5 ml micro centrifuge tube and 200 μl Buffer AE was pipetted
directly onto the DNeasy membrane, incubated at room temperature for 1 min and then
centrifuge for 1 min at 8000 rpm to elute.
VII.
Concentration and purity of DNA was
measured on Nanophotometer
16S rRNA
amplification
One of the most attractive potential uses of
16S rRNA gene sequence informatics is to provide genus and species identification
for isolates that do not fit any recognized biochemical profiles, for strains
generating only a “low likelihood” or “acceptable” identification [29,30].
Polymerase Chain Reaction (PCR) was performed using the Hi-Media Taq
polymerase, Hi-Media 50 mM MgCl2 and Hi-Media 10x buffer and QIAGEN
dNTPs (10 mM each). Universal 16S rRNA forward and reverse primer was used. PCR
amplifications were performed with an Applied Biosystems Veriti Thermal cycler.
The Master Mixture is presented in Table 2. The PCR protocol (conditions
which were used for thermal cycling) is shown in Table 3 [31]. The PCR product is stored at 4°C till infinity.
Sequencing of partial 16S rRNA gene
Purified PCR products samples were sent to
Macrogene Company, Rockville, Maryland, USA for further amplification in one
direction with the 16S primers using Big Dye Terminator Ready Reaction Mix
(ABI) and Sequencing of amplified product was done with the use of an ABI 3130
Genetic Analyzer version at Macrogene Company.
SEQUENCE ANALYSIS
Partial 16S rRNA gene sequence of studied
bacteria was analyzed with nucleotide BLAST search in GenBank from NCBI.
Phylogenetic relationship of this species was analyzed with other closely
related bacterial species present in GenBank.
Evolutionary
relationships of taxa and phylogenetic tree
The sequence generated a table of closely
similar organism with test organism. These similar organisms were selected and
there sequences were obtained in FASTA format. Once these sequences were
collected from BLAST, there sequences were checked for Multiple Sequence
Alignment (MSA) using T-Coffee tool from EBI (EMBL). The evolutionary history
was inferred using the Neighbor-Joining method [32]. The tree is drawn to
scale, with branch lengths in the same units as those of the evolutionary
distances used to infer the phylogenetic tree. The evolutionary distances were
computed using the number of differences method [33] and are in the units of
the number of base differences per sequence. Codon positions included were 1st+2nd+3rd+Noncoding.
All positions containing gaps and missing data were eliminated. Evolutionary
analyses were conducted in MEGA7 [34].
DATA ANALYSIS
Analysis of fish
growth performance
Fish growth performance was determined
illustrated by Agbebi et al. [35] in term of Final Individual Weight, Survival
(%), Specific Growth Rate (SGR%/day). The growth parameters were calculated at
the end of the experiment is:
Percentage weight gain PWG
(%) =
Specific growth rate, SGR =
where,
W1=Initial weight gained
W2=Final weight gained
Ln=Natural logarithm
Survival rate =
STATISTICAL ANALYSIS
Primary data obtained from the growth and
bacterial load were subjected to one way analysis of variance (ANOVA). Turkey’s
multiple comparisons test was used for comparison among diets means at a
significant level of 0.05. Computations were subjected to GraphPad Prism
version 7.04.
RESULTS
Growth performance
of fish
The growth performance of C. gariepinus fingerlings fed C. odorata leaves at three varying
levels of dietary supplementation is shown in Table 4. The highest final mean weight (1730.46 ± 87.27 g) recorded
in fish fed 1.5% C. odorata leaves
diets was significantly higher (p<0.05) than the lowest (711.63 ± 8.78 g) in
fish fed the control diet. The weight gain (1583.46 ± 59.33) was highest in
fish fed 1.5% C. odorata leaves diets
and the lowest (526.13 ± 9.26) was observed in fish fed the control diet.
Intestinal
microflora of the fish
Microbial count of
fish: The total
bacteria count and fungal counts in the intestine of fish fed C. odorata leaves diets and the control
is presented in Table 5. There was a
significant difference (p<0.05) in the total bacteria counts among fish C. odorata leaves paste and the control
having the lowest counts.
Biochemical
characterization of the bacteria isolate
Table 6 shows the biochemical test of the
bacteria isolated from the intestine of the fish fed C. odorata leaves and the control.
MOLECULAR CHARACTERIZATION OF BACTERIA
CULTURE ISOLATES
Sequencing of bacteria culture isolated from
intestine of fish fed the control diet
Partial 16S
gene of 536 bp was obtained after sequencing and shown below in FASTA format.
>TGCAAGTCGAACGGTGATCGCGCAGCAGCTTGCTGCTTCGCTGACGAGTGGCGGACGGGTGAGTAATGTCTGGGAAACTGCCTGATGGAGGGGGATAACTACTGGAAACGGTAGCTAACTACCGCATAACAGTCGCAAGACCAAAGAGGGGGACCTTCGGGCCTCTCTTGCCATCGGATGTGCCCAGATGGGATCTAGCTAGTAGGTGGGGTAACGGCTCACCTAGGTGCGACGATCCCTAGCTGGTCTGAGAGGATGACCAGGCCACACTGGAACTGAGACACGGTCCAGACTCCTACGGGAGGCAGCAGTGGGGAATATTGCACAATGGGCGCAAGCCTGATGCAGCCATGCCGCGTGTATGAAGAAGGCCTTCGGGTTGTAAAGTACTTTCAGCGGGGAGGAAGGGAGTAAAGTTAATACCTTTGCTCATTGACGTTACCCGCAGAAGAAGCACCGGCTAACTCCGTGCCAGCAGCCGCGGTAATACGGAGGGTGCAAGCGTTAATCGGAATTACTGGGCGTAAAGCGCACGGATGGTGA
Sequence analysis
Total 536
bp partial 16S rRNA sequence was retrieved in FASTA format and subjected for
BLAST search in GenBank. BLAST result showed that the test organism was similar
to Shigella flexneri (strain=SIA2)
with 97% similarity and E value 0.0.
Figure 1 presented the alignment of BLAST query and
the sequences producing significant alignment is shown in Figure 2.
Evolutionary relationships of taxa
Phylogenetic
relationship of test organism was analyzed with other partial 16S rRNA sequence
of similar microorganisms. The phylogenetic tree as shown in Figure 3 has the optimal tree with the
sum of branch length=41.81250000 as shown. The analysis involved 11 nucleotide
sequences; there were a total of 513 positions in the final dataset.
Sequencing of bacteria culture isolated from
intestine of fish fed the 0.5% C. odorata
leaves diet (first replicate)
Partial 16S
gene of 517 bp was obtained after sequencing and shown below in FASTA format.
>ACGTCTCGACGTGCAACGCGAAGAACCTTACCTGGTCTTGACATCCACAGAACTTTCCAGAGATGGATTGGTGCCTCTCGGGAACTGTGAGACAGGTGCTGCATGGCTGTCGTCAGCTCGTGTTGTGAAATGTTGGGTTAAGTCCCGCAACGAGCGCAACCCTTATCCTTTGTTGCCAGCGGTTAGGCCGGGAACTCAAAGGAGACTGCCAGTGATAAACTGGAGGAAGGTGGGGATGACGTCAAGTCATCATGGCCCCCTTACGACCAGGGCTACACACGTGCTACAATGGCATATACAAAGAGAAGCGACCTCGCGAGAGCAAGCGGACCTCATAGAAGTATGTCGTAGTCCGGATTGGAGTCTGCAACTCGACTCCATGAAGTCGGAATCGCTAGTAATCGTAGATCAGAATGCTACGGTGAATATCGTTCCCGGGCCTTGTACACACCGCCCGTCACACCATGGGAGTGGGTTGCAAAAGAAGTAGGTAGCTTAACCTTCGGGAGGGCYTTAGCCT
Sequence analysis
Total 517
bp partial 16S rRNA sequence was retrieved in FASTA format and subjected for
BLAST search in GenBank. BLAST result showed that the test organism was similar
to Klebsiella pneumonia (strain=SIF2)
with 99% similarity and E value 0.0.
Figure 4 presented the alignment of BLAST query and
the sequences producing significant alignment is shown in Figure 5.
Evolutionary relationships of taxa
Phylogenetic
relationship of test organism was analyzed with other partial 16S rRNA sequence
of similar microorganisms. The phylogenetic tree as shown in Figure 6 has the optimal tree with the
sum of branch length=2053.55468750 as shown. The analysis involved 17
nucleotide sequences; there were a total of 468 positions in the final dataset.
Sequencing of
bacteria culture isolated from intestine of fish fed the 0.5% C. odorata leaves diet (second
replicate)
Partial 16S gene of 401 bp was obtained after
sequencing and shown below in FASTA format.
>TCGGGAACTGTAAGACAGGTGCTCCATGGCGGTCTTCAGCTCGTGTTGTGAAATGTTGGGTTAAGTCCCGCAACGAGCGCAACCTTTTTCCTTTTTTCCCAGCGGTTAGGCCGGGAACTCAAAGGAGACTGCCAGTGATAAACTGGAGGAAGGTGGGGATGACGTCAAGTCATCATGGCCCTTACGACCAGGGCTACACACGTGCTACAATGGCAGATACAAAGAGAAGCGACCTCGCGAGAGCAAGCGGACCTCATAAAGTATGTCGTAGTCCGGATTGGAGTCTGCAACTCGACTCCATGAAGTCGGAATCGCTAGTAATCGTAGATCAGAATGCTACGGTGAATACGTTCCCGGGCCTTGTACACACCGCCCGTCACACCATGGGAGTGGGTTGCAA
Sequence analysis
Total 401 bp partial 16S rRNA sequence was
retrieved in FASTA format and subjected for BLAST search in GenBank. BLAST
result showed that the test organism was similar to Klebsiella pneumoniae (strain=SIG2) with 98% similarity and E value
0.0.
Figure 7 presented the alignment of BLAST
query and the sequences producing significant alignment is shown in Figure 8. The phylogenetic tree is
shown in Figure 6.
Sequencing of bacteria culture isolated from
intestine of fish fed the 1% C. odorata
leaves diet
Partial 16S
gene of 488 bp was obtained after sequencing and shown below in FASTA format.
AGCGGTAGCACAGAGAGCTTGCTCTCGGGTGACGAGCGGCGGACGGGTGAGTAATGTCTGGGAAACTGCCTGATGGAGGGGGATAACTACTGGAAACGGTAGCTAATACCGCATAACGTCGCCACAAGACCGCGAAAGTGGGGGACCTTCGGGCCTCATAGCCATCAGATGTGCCCAGATGTGGGATTAGCTAGTAGGTGGGGTAACGGCTCACCTAGGCGACGATCCCTAGCTGGTCTGAGAGGATGACCAGCCACACTGGAACTGAGACACGGTCCAGACTCCTACGGGAGGCAGCAGTGGGGAATATTGCACAATGGGCGCAAGCCTGATGCAGCCATGCCGCGTGTGTGAAGAAGGCCTTCGGGTTGTGCAAAGCACTTTCAGACGGGGAGGAAGGCGATAAGGTTAATAACCTCGTCGATTGACGTTACCCGCAGAAGAAGCACCGGCTAACTCCGTGCCAGCAGCCGCTGGTAATACTTGGA
Sequence analysis
Total 488
bp partial 16S rRNA sequence was retrieved in FASTA format and subjected for
BLAST search in GenBank. BLAST result showed that the test organism was similar
to Klebsiella pneumoniae (strain=SIH2)
with 97% similarity and E value 0.0.
Figure 9 presented the alignment of BLAST query and
the sequences producing significant alignment is shown in Figure 10.
Evolutionary relationships of taxa
Phylogenetic
relationship of test organism was analyzed with other partial 16S rRNA sequence
of similar microorganisms. The phylogenetic tree as shown in Figure 11 has the optimal tree with the
sum of branch length=283.94531250 as shown. The analysis involved 20 nucleotide
sequences; there were a total of 401 positions in the final dataset.
Sequencing of bacteria culture isolated from
intestine of fish fed the 1.5% C. odorata
leaves diet
Partial 16S
gene of 579 bp was obtained after sequencing and shown below in FASTA format.
>GAGCGGTAGCACAGAGAGCTTGCTCTCGGGTGACGAGCGGCGGACGGGTGAGTAATGTCTGGGAAACTGCCTGATGGAGGGGGATAACTACTGGAAACGGTAGCTAATACCGCATAATGTCGCAAGACCAAAGTGGGGGACCTTCGGGCCTCATGCCATCAGATGTGCCCAGATGGGATTAGCTAGTAGGTGGGGTAACGGCTCACCTAGGCGACGATCCCTAGCTGGTCTGAGAGGATGACCAGCCACACTGGAACTGAGACACGGTCCAGACTCCTACGGGAGGCAGCAGTGGGGAATATTGCACAATGGGCGCAAGCCTGATGCAGCCATGCCGCGTGTGTGAAGAAGGCCTTCGGGTTGTAAAGCACTTTCAGCGGGGAGGAAGGCGTTGAGGTTAATAACCTTGGCGATTGACGTTACCCGCAGAAGAAGCACCGGCTAACTCCGTGCCAGCAGCCGCGGTAATACGGAGGGTGCAAGCGTTAATCGGAATTACTGGGCGTAAAGCGCACGCAGGCGGTCTGTCAAGTCGGATGTGAAATCCCCGGGCTCAACCTGGGAACTGCATTCGAAACTGGCAGGCTAGAGTCTTGTAGA
Sequence analysis
Total 579
bp partial 16S rRNA sequence was retrieved in FASTA format and subjected for
BLAST search in GenBank. BLAST result showed that the test organism was similar
to Klebsiella pneumoniae
(strain=SIA3) with 99% similarity and E value 0.0.
Figure 12 presented the alignment of BLAST query and the sequences producing significant alignment is shown in Figure 13.
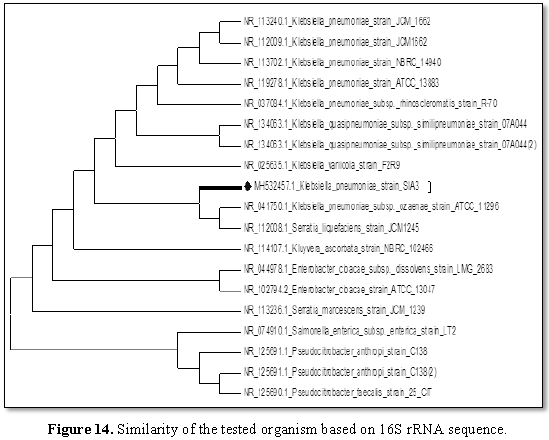
Evolutionary relationships of taxa
Phylogenetic relationship of test organism was analyzed with other partial 16S rRNA sequence of similar microorganisms. The phylogenetic tree as shown in Figure 14 has the optimal tree with the sum of branch length=59.31250000 as shown. The analysis involved 19 nucleotide sequences; there were a total of 588 positions in the final dataset.The summary of bacterial species identified is shown in Table 7.
DISCUSSION
This study has proven that molecular identification is a promising and better approach in identification of microorganism through their DNA. The biochemical identification revealed the presence of Micrococcus sp. in the fish intestine fed control diet while the molecular method revealed S. flexneri. Similar variation was observed in fish intestine fed 1% C. odorata leaves as biochemical identification revealed Shigella sp. while molecular revealed K. pneumonia.
Information
in this present study has revealed the antibacterial potential of supplementing
the leaves of C. odorata as it
inhibited the growth of some enterobacteriaceae (K. pneumonia and Proteus
mirabilis) found in the intestine of C.
gariepinus. The report of Vital and Rivera [36] corroborated this present
study in that ethanolic extract of C.
odorata leaves showed antimicrobial activity against Bacillus subtilis, Staphylococcus
aureus and S. typhimurium through
inhibiting the growth of these bacteria when challenged with C. odorata leaves extract and the author
suggested that the ability of the leaves to inhibit growth of bacteria could be
due to presence of flavonoid and tannin found in the phytochemical analysis of
the leaves.
The DNA
sequencing analysis of the bacterial isolates of fish supplemented with C. odorata leaves showed the presence of
disease causing enterobacteriaceae but their effects are minimized as indicated
in the total bacteria counts of the fish intestine. This could infer that C. odorata leaves inhibited the growth
of these microorganisms and thus promoting the growth and health of the fish
fed the leaves. The highest weight gain, specific growth rate and survival rate
and lowest feed conversion ratio were recorded in fish fed the highest
inclusion level of C. odorata leaves.
This indicated that nutrient were better utilized in diet supplemented with C. odorata leaves and consequently the
dietary inclusion of C. odorata leaves
inhibited the growth of disease causing pathogen as seen in the gene sequencing
analysis. This remarkable achievement is due to the presence of bioactive
compounds imbedded in the leaves of C.
odorata. Several studies have proven that flavonoid, steroid and phenolic
compounds are linked to the growth promoting effect and antibacterial potential
of the leaves [11-13].
CONCLUSION
This
present study has revealed the effect of supplemented diets on the intestinal
bacteria of cultured fish based on the results obtained from gene sequence
analysis, which is a promising area in fish nutrigenomics and nutrigenetics. C. odorata leaves, most especially 1.5%
inclusion level have proven to have a significant effect in that it promotes
the growth of cultured C. gariepinus,
reduce the microbial load of the fish and subsequently inhibiting the growth of
enterobactriaceae found in the fish intestine through the gene sequencing.
ACKNOWLEDGEMENT
The author
thanked the efforts of the technologist, Mr. Omonigbeyin in the department of
Molecular biology, Covenant University, Ota, Ogun state during the DNA extraction
process. The assistance of Mrs. Adekeye of Department of Microbiology, Covenant
University is appreciated.
1.
Mukherjee PK, Wahile A (2006) Integrated approaches
towards drug development from Ayurveda and other Indian system of medicine. J
Ethanopharmacol 103: 5-35.
2.
Sliva Junior AA, Vizotto VJ, Giorgi E, Macedo SG, Marques
LF (1994) Plants medicinals, caracterizacao e cultivo EPAGRI. Bol Tecnico
Florianopolis 68: 1-71.
3.
Di Stasi LC (1996) Arte, ciencia e magia. In LC Di Stati,
CA Hiruma-Lima (eds), plantas Medicinais: Arte e Ciencia, Unesp, Sao Paulo, pp:
15-21.
4.
Odunbaku OA, Ilusanya OA (2008) Antibacterial activity of
the ethanolic and methanolic leaf extracts of some tropical plants on some
human pathogenic microbes. Res J Agric Biol Sci 4: 373-376.
5.
Hostettmann K, Marston A, Wolfender JL (2000) The
potential of African medicinal plants as a source of drugs. Curr Org Chem, pp:
2-48.
6.
Schmelzer GH, Gurib-Fakin A (2008) Medicinal Plants 1,
PROTA Foundation Netherlands/Backhuys Publishers, Leiden, Netherlands, p: 791.
7.
Ekenyem BU, Obih TKO, Odo, BI, Mba FIA (2010) Performance
of finisher broiler chicks fed varying replacement levels of Chromolaena odorata leaf for soyabean
meal. Pak J Nutr 9: 558-561.
8.
Akinmoladun AC, Ibokun EO, Dan-Ologe IA (2007) Phytochemical
constituents and antioxidant properties of extract from the leaves of Chromolaena odorata. Sci Res Essay 2:
191-194.
9.
Volk WA, Wheeler (1993) Basic Microbiology Miscellaneous:
Markham Edition V publisher. Jakarta, p: 390.
10.
Cowan MM (1999) Plants product as antimicrobial agent.
Clin Microbiol Rev 12: 564-582.
11.
Pelczar C (2005) Dasar-dasar Mikrobiologi I. (Hadioetomo
RS, Imas T, Tjitrosomo SS, Angka SL. Jakarta, Trans.). Penerbit Universitas
Indonesia, p: 508.
12.
Mori A, Nishino C, Enoki N, Tawata S (1987) Antibacterial
activity and mode of action of plant flavonoids against Proteus vulgaris and Staphylococcus
auratus. Phytochemistry 26: 2231-2234.
13.
Putra INK (2007) Study power antimicrobial preservatives
plant extract multiple materials destroyer Nira nira against microbes and
gynecology actively compounds. (Unpublished Doctoral Dissertation). University
of Brawijaya, Malang.
14.
Ghormade V, Khare A, Baghel RPS (2011) Nutrigenomics and
its applications in animal science. Vet Res Forum 2: 147-155.
15.
Banerjee G, Pal R, Ray AK (2015) Applications of
nutrigenomics in animal sectors: A review. Asian J Anim Vet Adv 10: 489-499.
16.
Harland JI (2005) Nutrition and genetics, mapping
individual health.
17.
Garg R, Sharma N, Jain SK (2014) Nutrigenomics and
nutrigenetics: Concepts and applications in nutrition research and practice.
Acta Medica Int 1: 124-130.
18.
Ordovas JM, Mooser V (2004) Nutrigenomics and
nutrigenetics. Curr Opin Lipidol 15: 101-108.
19.
Tellez G, Latorre JD, Kuttappan VA, Kogut MH, Wolfenden A
(2014) Utilization of rye as energy source affects bacterial translocation,
intestinal viscosity, microbiota composition and bone mineralization in broiler
chickens. Front Genet 5: 339.
20.
Costa NMB, Rosa COB (2011) Functional foods: Bioactive
components and physiological effects. 1 Reprint, R´ubio, Rio de Janeiro,
Brazil.
21.
Morozova O, Marra MA (2008) Applications of
next-generation sequencing technologies in functional genomics. Genomics 92:
255-264.
22.
Ogawa G, Ishida, M, Kato H, Fujise Y, Urano N (2010)
Identification of facultative anaerobic bacteria isolated from the intestine of
the minke whale Balaenoptera acutorostrata by 16S rRNA sequencing analysis.
Fisheries Sci 76: 177-181.
23.
Kalu BA, Njike MC, Ikurior SA (1986a) Evaluating the
potential of Tridax procumbens for
livestock feed. I. Morphological stages of development and chemical
composition. Nig J Anim Prod 13: 11-12.
24.
Ajibola SI, Obasa, SO, Akintokun AK, Abdulraheem I (2016)
Body weight changes, nutrient utilization and intestinal microflora of African
catfish (Clarias gariepinus) fed Aloe barbadensis leaves. Nig J Anim Prod
43: 385-398.
25.
Chessbrough M (2002) District laboratory practice in
tropical countries. Part 1. Cambridge low price edition. Cambridge University
Press, London.
26.
Cowan ST, Steel KJ (1993) Enterobacteriaceae, in Barrow,
G.I and. Felthan, R. K. A (Eds). Manual for the Identification of Medical
Bacteria (3rd
Edn),
Cambridge University press, United Kingdom, pp: 213-218.
27.
Brahmbhatt DN (2012) Molecular identification of bacteria
using 16S rDNA sequencing. Masters Dissertation, Gujarat University,
Ahmeddabad, India, pp: 1-50.
28.
Corkill G, Raphley R (2008) The manipulation of nucleic
acids: Basic tools and techniques, Edited by Walker J and Raphley R. In:
Molecular Biomethods Handbook, 2nd Edn. Humana Press, pp: 3-15.
29.
Drancourt M, Bollet C, Carlioz A, Martelin R, Gayral J,
et al. (2000) 16S ribosomal DNA sequence analysis of a large collection of
environmental and clinical unidentifiable bacterial isolates. J Clin Microbiol
38: 3623-3630.
30.
Mignard S, Flandrois J (2006) 16S rRNA sequencing in
routine bacterial identification: A 30 month experiment. J Microbiol Methods
67: 574-581.
31.
Ausubel FM (1991) Current protocols in molecular biology.
John Wiley and Sons, Inc. New York.
32.
Saitou N, Nei M (1987) The neighbor-joining method: A new
method for reconstructing phylogenetic trees. Mol Biol Evolution 4: 406-425.
33.
Nei M, Kumar S (2000) Molecular evolution and
phylogenetics. Oxford University Press, New York.
34.
Kumar S, Stecher G, Tamura K (2016) MEGA7: Molecular
evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol
33: 1870-1874.
35.
Agbebi OT, Lawal HB, Odebiyi VC (2012) Aflatoxin effect
of moulded gel waste mixed with ginger and its histopathological study on Clarias garepinus. Glob J Sci Front Res
12: 7-15.
36.
Vital PG, Rivera WL (2009) Antimicrobial activity and
cytoxicity of Chromolaena odorata (L)
and Uncaria perrottetii (A. Rich)
extracts. J Med Plants Res 3: 511-518.