826
Views & Citations10
Likes & Shares
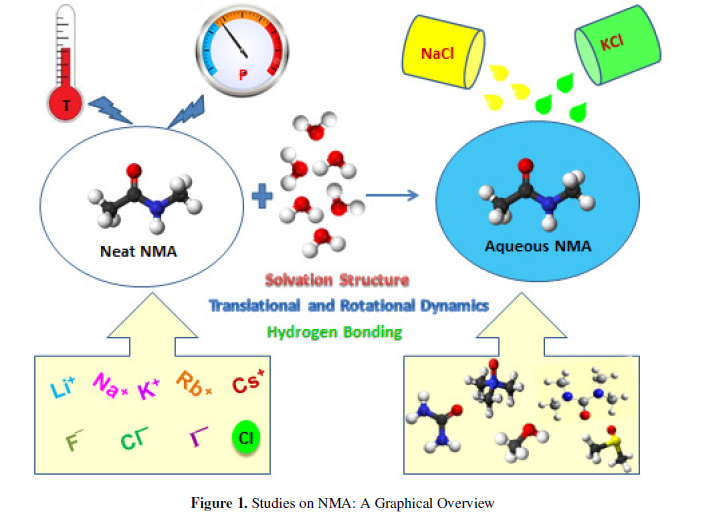
In this mini review, we report a brief overview of results of molecular dynamics simulations of N-methylacetamide and its aqueous solution at different thermodynamic conditions. The structure and dynamical properties of neat NMA are analysed at different temperatures and pressures. The results are highlighted in terms of various dynamical properties associated with translational and rotational motion of neat NMA. The results of solvation structure and dynamics of different alkali metal and halide ions in liquid NMA are also mentioned. Then we proceed for discussing the behaviour of NMA and its aqueous solutions with varying salt concentration. How the structure and dynamics of aqueous NMA solution are changing in presence of different cosolvents are also highlighted. Finally, the recent study of the influence of dimethyl sulfoxide, ethanol and trifluoroethanol on hydrogen bonding behaviour of aqueous NMA system is discussed.
Keywords: Molecular dynamics simulation, N-methyacetamide, Hydrogen bond dynamics.
Also, they noticed that on applying pressure,
peak heights for NMA-NMA site-site correlations were reduced. They contrasted
these results to our work, suggesting that the overall change in surrounding
water molecules may alter how pressure affects NMA-NMA association.
In biologically active systems, peptide
linkages are often immersed in aqueous system with presence of several ions.
Keeping this concept in mind, we designed a study where we could zoom on to
subtle interplays of hydrogen bonding along with ion coordination and dynamics
behaviour in aqueous environment of peptide bonds [9]. Theoretical and
experimental investigations on interactions of NMA with water were available in
literature [10-18]. A majority of the studies were devoted to influence of
aqueous environment changes on amide-I vibrations of N-methylacetamide [19-24].
In our study, Intra- and inter-molecular hydrogen bonds were
calculated between NMA-water with the help of certain geometric criteria
[25-31] where it is assumed that a hydrogen bond between two species exists, if
the following distance and angular criteria are satisfied, i.e., R(OX) < Rc(OX), R(OH)
< Rc(OH), and θ < θc. For example, R(OX)
and R(OH) denote the oxygen (water)-nitrogen (NMA) and oxygen
(water)-hydrogen(NMA) distances, and the corresponding quantities with
subscript “c” denote the cutoff values. The angle θ (=θ(NOH)) is the
nitrogen-oxygen-hydrogen angle, and the θc is the upper limit that
is allowed for hydrogen bond to exist between oxygen of water and the hydrogen
of the NMA molecule. The cutoff values for O-N and O-H distances are determined
from the positions of the first minimum of the corresponding radial
distribution functions. We have maintained the cutoff angle to be 45o
instead of giving 30o to allow for flexibility induced by thermal
effects. It was found that with dilution, though stability of both kinds of
hydrogen bonds decrease, average number of inter-molecular
NMA-water hydrogen bonds are found to increase in contrast to decrease of
NMA-NMA intra-molecular hydrogen
bonds. Even when water intra-molecular
hydrogen bonding becomes prevalent at higher dilution range, even then
stability of NMA-water hydrogen bonds continues to dominate. Hydrogen bond
energies for >C=ONMA...HWAT and N-HNMA...OWAT
hydrogen bonds have been calculated by Sarma and Paul [8] to be nearly
similar and they have also commented that this is consonant with the results of
our study. Recently, Yadav and Chandra [32] observed slower vibrational
spectral diffusion of water molecules in solvation shells of N-methylacetamide
and also proposed that average lifetime and residence time for ONMA...HWAT
hydrogen bonds were higher than that for HNMA...OWAT
hydrogen bonds.
In the same study Pattanayak and Chowdhuri [9],
considered biologically important ions (Na+, K+, Cl-)
and a neutral solute modelled by Cl were considered for the study and it was
revealed that while the positive and negative ions were strongly solvated by
pure NMA, as water is gradually introduced into the system, they had higher
inclination to keep water in their neighbourhood. On the other hand, the
neutral solute showed more affinity towards NMA due to its ability to be
solvated by NMA’s methyl groups. Strong solvation shells of the ionic solutes
made the translational dynamics slower for them as compared to faster motion of
the neutral solute. Around the same time , Algaey and Vegt compared
interactions of aqueous salt solutions with N-isopropylacrylamide (NiPAM) to
that with NMA. While NMA formed hydrogen bonds with all Hofmeister anions,
NiPAM did not show such hydrogen bonding interactions [33]. Encouraged by these
results, several simulations were conducted by Pattanayak and Chowdhuri to
explore behaviour of five alkali metal and halide ions in liquid NMA at two
different temperatures [34]. Smaller sized ions had stronger solvation shells
while increasing ion size led to monotonic increase in coordination number
which was further increased for lower temperature. At higher temperatures, less
structuring of solvation shells were noted and the effect was more apparent for
anionic moieties where interactions with NMA were disrupted. Anomalous ion size
dependence of diffusion coefficients were observed which were attributed to
high dielectric friction in NMA and also for neutral solutes, higher Stokes
friction was postulated to be a factor in the diffusion being less in NMA than
for water or methanol. Beck et al. studied small clusters formed of an ion,
N-methylacetamide and incrementing number of water molecules of the form
Cl(NMA)1(H2O)0–2Ar2 , through Infrared
Predissociation (IRPD) spectra combined with Born–Oppenheimer Molecular
Dynamics (BOMD) IR spectra . It was demonstrated how Cl– forms a
strong ionic hydrogen bond to the H-N(NMA), due to which the N-H stretch is
weakened and shifted to lower frequency [35] .
The next step to enhance our understanding of
behaviour of solutes in presence of NMA was to observe structure and dynamics
of salt solutions in liquid as well as aqueous solution of NMA. Initially,
different concentrations of NaCl were taken to observe variation of hydrogen
bonding structure in liquid NMA [36]. From the amide-amide radial distribution
functions, hydrogen bond energy and number variation trends, it was clear that
with increasing salt concentration, hydrogen bonding in NMA was perturbed and destabilised.
Contact-ion pair influences were evident when ion-NMA site-site correlations
were investigated and residence times were found to be higher for sodium ion
near NMA, relative to the anion. Friction due to ion atmosphere was regarded as
the main reason behind slowing of translational as well as rotational dynamics
of NMA as well as the ions.
Subsequently, effects of NaCl and KCl were
explored in aqueous solutions of NMA which more realistically mimic the
biological and cellular environments [37]. Previously, Jungwirth and co-workers
[38] had investigated solvation shell of ions in aqueous NMA solutions
containing 1M concentration of various salt solutions. We found that with
increasing salt concentration, NaCl enhances ion interactions with NMA and
water at the cost of NMAs interactions with itself and water while the opposite
effect was seen in case of KCl, which had stronger contact-pair formation
probability. Slower translational and rotational dynamics as well as structural
relaxation times of NMA-water hydrogen bonds were more noticeable in
concentrated NaCl in comparison to KCl solution.
Having concluded such studies on behaviour of
liquid as well as aqueous NMA solutions in presence of ion and salt
concentrations, we shifted our focus to various cosolvents and cosolutes which
may surround the vicinity of aqueous environment of peptide linkages. We
started our journey in this area [39] by
considering system of NMA in methanol-water solution. Methanol was chosen owing
to its wide range of biological activity including structural effects on
proteins; its extensive hydrogen bonding network in water as well as
experimental evidences of effects on NMA [40,41]. It was found that addition of
even high concentration of methanol could not alter the preference of NMA to
hydrogen bond with water than with methanol, as was evident from strong
interaction energies and stable NMA-water hydrogen bonds. While water oxygen
showed preference to stay in vicinity of methyl group of NMA, methanol strongly
interacted with the non-polar sites of NMA with its methyl groups instead.
We then conducted a comparative investigation
of effects of important cosolutes like urea, trimethylamine N-oxide (TMAO) and
tetramethylurea (TMU) on behaviour of aqueous NMA solution [42]. These
co-solutes form part of a special class of molecules known as osmolytes which
protect proteins against environmental stresses and also affect their
folding/conformational equilibria [43-50]. Earlier, several experimental
studies had been conducted on the structural dynamics of water in presence of
such osmolytes [51-53]. Gao et al set precedence by exploring solution
structure of specific protein backbones in presence of these osmolytes [54]. In
our study, we found that NMA-NMA hydrogen bonding tendency were enhanced by
TMAO and TMU relative to urea and number of water molecules near methyl surface
of NMA were significantly reduced in presence of TMU. An interesting
observation came through, that NMA partners with TMU to give rise to water
self-segregation but NMA-water hydrogen bonds were more stable in TMU solution.
Urea affected NMA’s tendency to donate hydrogen to water oxygen but did not
have any appreciable effects on hydrogen bonding structure and dynamics of
aqueous NMA solution. Since the molecule of TMAO could be described by three
force field models i.e Kast, Garcia and Netz, we [55] carried out a separate
study to compare them in their relative interactions with aqueous NMA solution.
Compared to the widely used Kast model, Garcia and Netz model increased
hydrogen bonding with NMA amide hydrogen with oxygen of TMAO. Particularly,
Netz model decreased HNMA...OWAT hydrogen bonding
interactions owing to affinity of TMAO oxygen towards amide site of NMA.
Three-hydrogen bonded water complexes of TMAO were more favoured by Garcia and
Netz models in comparison with Kast model.
Paul and Paul [56,57] studied the effects of trehalose on aqueous solvation
of NMA and also investigated its role in counteracting effects of urea on
hydrogen bonding in N-methylacetamide solution. It was observed that water-NMA
hydrogen bonds were gradually replaced by NMA-trehalose hydrogen bonds and
NMA-urea hydrogen bonds in their binary solutions but in ternary solutions,
trehalose molecules were preferred to be in vicinity of NMA surfaces than urea
and a decrease in interaction energy between NMA-urea molecules was seen in
ternary urea-trehalose-NMA system.
Recently, influence of DMSO on hydrogen bonding
behaviour of aqueous NMA system was studied by Chand and Chowdhuri [58] owing
to diverse range of effects that DMSO has on proteins. In aqueous environment,
NMA forms the strongest hydrogen bonds to DMSO than to water or itself with
orientation of DMSO molecule towards amide hydrogen site of NMA. At high DMSO
concentrations, lower lifetimes of HNMA...OWAT hydrogen
bonds were noted, where the stability of HNMA...ODMSO
hydrogen bond was found to increase. Methyl-methyl interaction of DMSO and NMA
molecules reduces but presence of high DMSO concentrations dehydrates nearby
water molecules from the methyl site of NMA. In a recent study, Chand and
Chowdhuri [59] have also examined the behaviour of ethanol and trifluoroethanol
on hydrogen bonding properties of aqueous N-methylacetamide. It is observed
that ethanol favours aqueous solvation of N-methylacetamide but
trifluoroethanol maintains NMA-NMA hydrogen bonding while also donating
hydrogen bond to carbonyl oxygen of NMA.
This review highlights some recent
computational investigations on the structure and hydrogen bond dynamics of NMA
and its aqueous solution. Presently, our endeavours are directed at more such
explorations of influence of mixtures of such co-solutes on aqueous environment
of N-methylacetamide in an effort to bring us one step closer to the
microscopic investigations of the peptide bond and the hydrogen bonding
networks and dynamics therein, which will bring clarity in comprehension of
protein-solvent interactions in future.
Acknowledgment
Authors are grateful to the Department of
Science and Technology (DST), Government of India, for the financial support to
this work through Grant No. SB/S1/PC- 28/2012; Council of Scientific and
Industrial Research (CSIR), Government of India for SRF-fellowship and also to
the Indian Institute of Technology, Bhubaneswar for all kinds of support to
execute the Project. SKP would like to acknowledge to National Institute of
Technology Raipur for support.
- Klotz IM, Franzen JS (1962) Hydrogen bonds between model peptide groups in solution. J Am Chem Soc 84: 3461-3466.
- MacKerell Jr AD, Bashford D, Bellott M, Dunbrack Jr RL, Evanseck JD, et al. (1998) All-atom empirical potential for molecular modeling and dynamics studies of proteins. J Phys Chem B 102 18: 3586-3616.
- Jorgensen WL, Swenson CJ (1985) Optimized intermolecular potential functions for amides and peptides. Structure and properties of liquid amides. J Am Chem Soc 107: 569-578.
- Katz JL, Post B (1960) The crystal structure and polymorphism of N-methylacetamide. Acta Crystallographica 13: 624-628.
- Kitano M, Fukuyama T, Kuchitsu K (1973) Molecular structure of N-methylacetamide as studied by gas electron diffraction. Bull Chem Soc Japan 46: 384-387.
- Leader GR, Gormley JF (1951) The dielectric constant of N-methylamides. J American Chem Soc 73: 5731-5733.
- Pattanayak SK, Prashar N, Chowdhuri S (2011) Effect of temperature and pressure on the structure, dynamics, and hydrogen bond properties of liquid N-methylacetamide: A molecular dynamics study. J Chem Phys 134.
- Sarma R, Paul S (2012) Effect of pressure on the solution structure and hydrogen bond properties of aqueous N-methylacetamide. Chem Phys 407: 115-123.
- Pattanayak SK, Chowdhuri S (2011) Effect of Water on Solvation Structure and Dynamics of Ions in the Peptide Bond Environment: Importance of Hydrogen Bonding and Dynamics of the Solvents. J Phys Chem B 115: 13241-13252.
- Akiyama M (2002) Study on hydration enthalpy of N-methylacetamide in water. Spectrochim Acta Mol Biomol Spectrosc 58: 1943-1950.
- Allison SK, Bates SP, Crain J, Martyna GJ (2006) Solution structure of the aqueous model peptide N-methylacetamide. J Phys Chem B 110: 21319-21326.
- Besley NA, Hirst JD (1998) Ab initio study of the effect of solvation on the electronic spectra of formamide and N-methylacetamide. J Phys Chem A 102: 10791-10797.
- Gao J, Freindorf M (1997) Hybrid ab initio QM/MM simulation of N-methylacetamide in aqueous solution. J Phys Chem A 101: 3182-3188.
- Huelsekopf M, Ludwig R (2001) Correlations between structural, NMR and IR spectroscopic properties of N-methylacetamide. Magn Reson Chem 39: S127-S134.
- Lin B, Lopes PEM, Roux B, MacKerell AD Jr (2013) Kirkwood-Buff analysis of aqueous N-methylacetamide and acetamide solutions modeled by the CHARMM additive and Drude polarizable force fields. J Chem Phys 139.
- Luque F, Orozco M (1993) Theoretical study of N-methylacetamide in vacuum and aqueous solution: implications for the peptide bond isomerization. J Org Chem 58: 6397-6405.
- Yu HA, Karplus M, Pettitt BM (1991) Aqueous solvation of n-methylacetamide conformers: Comparison of simulations and integral equation theories. J Am Chem Soc 113: 2425-2434.
- Markham LM, Hudson BS (1996) Ab initio analysis of the effects of aqueous solvation on the resonance Raman intensities of N-methylacetamide. J Phys Chem 100: 2731-2737.
- Cazade PA, Hédin F, Xu ZH, Meuwly M (2015) Vibrational Relaxation and Energy Migration of N-Methylacetamide in Water: The Role of Nonbonded Interactions. J Phys Chem B 119: 3112-3122.
- De Camp M, DeFlores L, McCracken J, Tokmakoff A, Kwac K (2005) Amide I vibrational dynamics of N-methylacetamide in polar solvents: The role of electrostatic interactions. J Phys Chem B 109: 11016-11026.
- Nandini G, Sathyanarayana DN (2002) Ab initio studies on geometry and vibrational spectra of N-methyl formamide and N-methylacetamide. J Mol Struct: THEOCHEM 579: 1-9.
- Torii H, Tatsumi T, Kanazawa T, Tasumi, M (1998) Effects of intermolecular hydrogen-bonding interactions on the amide I mode of N-methylacetamide: matrix-isolation infrared studies and ab initio molecular orbital calculations. J Phys Chem B 102: 309-314.
- Chen X, Schweitzer-Stenner R, Krimm S, Mirkin NG, Asher SA (1994) N-methylacetamide and its hydrogen-bonded water molecules are vibrationally coupled. J Am Chem Soc 116: 11141-11142.
- Kim JH, Cho M (2003) Interplay of the intramolecular water vibrations and hydrogen bond in N-methylacetamide-water complexes: Ab initio calculation studies. Bull Korean Chem Soc 24: 1061-1068.
- Chowdhuri S, Chandra A (2006) Dynamics of halide ion-water hydrogen bonds in aqueous solutions: Dependence on ion size and temperature. J Phys Chem B 110: 9674-9680.
- Chandra A (2000) Effects of ion atmosphere on hydrogen-bond dynamics in aqueous electrolyte solutions. Phys Rev Lett 85: 768.
- Chowdhuri S, Chandra A (2003) Pressure effects on the tracer diffusion and orientational relaxation of hydrogen bonding solutes in ambient and supercooled water. Chem Phys Letters 373: 79-86.
- Chandra A, Chowdhuri, S (2002) Pressure effects on the dynamics and hydrogen bond properties of aqueous electrolyte solutions: The role of ion screening. J Phys Chem B 106: 6779-6783.
- Luzar A, Chandler D (1996) Hydrogen-bond kinetics in liquid water. Nature (London) 379: 55-57.
- Kumar R, Schmidt J, Skinner J (2007) Hydrogen bonding definitions and dynamics in liquid water. J Chem Phys 126: 204107.
- Xu H, Berne B (2001) Hydrogen-bond kinetics in the solvation shell of a polypeptide. J Phys Chem B 105: 11929-11932.
- Yadav VK, Chandra A (2015) First-Principles Simulation Study of Vibrational Spectral Diffusion and Hydrogen Bond Fluctuations in Aqueous Solution of N-Methylacetamide. J Phys Chem B 119: 9858-9867.
- Algaer EA, van der Vegt NF (2011) Hofmeister ion interactions with model amide compounds. J Phys Chem B 115: 13781-13787.
- Pattanayak SK, Chowdhuri S (2012) Size Dependence of Solvation Structure and Dynamics of Ions in Liquid N-methylacetamide: A Molecular Dynamics Simulation Study. J Theor Comput Chem 11: 361-377.
- Beck JP, Gaigeot MP, Lisy JM (2013) Anharmonic vibrations of N–H in Cl−(N-methylacetamide) 1 (H 2 O) 0–2 Ar 2 cluster ions. Combined IRPD experiments and BOMD simulations. Phys Chem Chem Phys 15: 16736-16745.
- Pattanayak SK, Chowdhuri S (2012) A molecular dynamics simulations study on the behavior of liquid N-methylacetamide in presence of NaCl: Structure, dynamics and H-bond properties. J Mol Liq 172: 102-109.
- Pattanayak SK, Chowdhuri S (2013) Effects of concentrated NaCl and KCl solutions on the behaviour of aqueous peptide bond environment: single-particle dynamics and H-bond structural relaxation. Mol Phys 111: 3297-3310.
- Heyda J, Vincent JC, Tobias DJ, Dzubiella J, Jungwirth P (2009) Ion specificity at the peptide bond: Molecular dynamics simulations of N-methylacetamide in aqueous salt solutions. J Phys Chem B 114: 1213-1220.
- Pattanayak SK, Chowdhuri S (2014) Effects of methanol on the hydrogen bonding structure and dynamics in aqueous N-methylacetamide solution. J Mol Liq 194: 141-148.
- Woutersen S, Mu Y, Stock G, Hamm P (2001) Hydrogen-bond lifetime measured by time-resolved 2D-IR spectroscopy: N-methylacetamide in methanol. Chem Phys 266: 137-147.
- Kwac K, Cho M (2005) Hydrogen bonding dynamics and two‐dimensional vibrational spectroscopy: N‐methylacetamide in liquid methanol. J Raman Spectrosc 36: 326-336.
- Pattanayak SK, Chettiyankandy P, Chowdhuri S (2014) Effects of co-solutes on the hydrogen bonding structure and dynamics in aqueous N-methylacetamide solution: a molecular dynamics simulations study. Mol Phys 112: 2906-2919.
- AJ W, DW B (1997) A naturally occurring protective system in urea-rich cells: mechanism of osmolyte protection of proteins against urea denaturation. Biochem 36: 9101.
- Auton M, Baskakov I, Bolen CL, Bolen DW (2001) On identifying the fundamental forces of osmolyte-induced protein stability. Biophys J 80: 558A-558A.
- Auton M, Bolen DW (2005) Predicting the energetics of osmolyte-induced protein folding/unfolding. Proc Natl Acad Sci USA 102: 15065-15068.
- Auton M, Bolen DW, Rosgen J (2008) Structural thermodynamics of protein preferential solvation: Osmolyte solvation of proteins, aminoacids, and peptides. Proteins: Struct Funct Bioinf 73: 802-813.
- Bruzdziak P, Panuszko A, Stangret J (2013) Influence of Osmolytes on Protein and Water Structure: A Step to Understanding the Mechanism of Protein Stabilization. J Phys Chem B 117: 11502-11508.
- Canchi DR, Garcia AE (2013) Cosolvent Effects on Protein Stability. Annu Rev Phys Chem 64 : 273-293.
- Khan SH, Ahmad N, Ahmad F, Kumar R (2010) Naturally occurring organic osmolytes: From cell physiology to disease prevention. IUBMB Life 62: 891-895.
- Rosgen J, Pettitt BM, Bolen DW (2007) An analysis of the molecular origin of osmolyte-dependent protein stability. Protein Sci 16: 733-743.
- Rezus YLA, Bakker HJ (2009) Destabilization of the Hydrogen-Bond Structure of Water by the Osmolyte Trimethylamine N-Oxide. J Phys Chem B 113: 4038-4044.
- Rezus YLA, Bakker HJ (2006) Effect of urea on the structural dynamics of water. Proc Natl Acad Sci USA 103: 18417-18420.
- Kuffel A, Zielkiewicz J (2010) The hydrogen bond network structure within the hydration shell around simple osmolytes: Urea, tetramethylurea, and trimethylamine-N-oxide, investigated using both a fixed charge and a polarizable water model. J Chem Chem Phys 133: 035102.
- Wei H, Fan Y, Gao YQ (2009) Effects of urea, tetramethyl urea, and trimethylamine N-oxide on aqueous solution structure and solvation of protein backbones: a molecular dynamics simulation study. J Phys Chem B 114: 557-568.
- Chand A, Chettiyankandy P, Pattanayak SK, Chowdhuri S (2016) Effects of trimethylamine-N-oxide (TMAO) on aqueous N-methylacetamide solution: A comparison of different force fields of TMAO. J Mol Liq.
- Paul S, Paul S (2014) Trehalose Induced Modifications in the Solvation Pattern of N-Methylacetamide. J Phys Chem B 118: 1052-1063.
- Paul S, Paul S (2015) Exploring the Counteracting Mechanism of Trehalose on Urea Conferred Protein Denaturation: A Molecular Dynamics Simulation Study. J Phys Chem B 119: 9820-9834.
- Chand A, Chowdhuri S (2016) Effects of dimethyl sulfoxide on the hydrogen bonding structure and dynamics of aqueous N-methylacetamide solution. J Chem Sci 128: 991-1001.
- Chand A, Chowdhuri S (2016) Behaviour of aqueous N-methylacetamide solution in presence of ethanol and 2, 2, 2 tri-fluoroethanol: Hydrogen bonding structure and dynamics. J Mol Liq.