1353
Views & Citations353
Likes & Shares
This
pilot study investigated 73 healthy and diseased periodontal pockets evaluated
by micropipette analysis to evaluate the bacteria existing in healthy and
diseased pockets. The biofilm in the pockets are compared to 16 swish and 14
irrigation and swish saliva samples. Comparing the oral saliva samples (swish
and irrigation/swish) pocket biofilm results to micropipette analysis of the
healthy and diseased periodontal pockets enables an evaluation of consistency.
Noteworthy differences were found. The patients came from two separate offices
(one periodontist and one general dentist) and the evaluators were calibrated
for reliability.
The
patients were categorized as free of periodontal disease (PD) having 3 mm
pockets or less without bleeding upon probing (BOP) and patients with
periodontal disease having 3 mm pockets with BOP or 4 mm pockets or greater.
Bacteria were collected by micropipette from the periodontal sulcus or pocket
and were evaluated by DNA analysis (MicroGenDx) by pocket depth to the genus/species
level. The bacterial classification compared type of bacteria by a response to
Gram stain (Gram-positive or Gram-negative) and categorized the bacteria as;
anaerobic, facultative anaerobes and aerobic. Fungi were also evaluated. A log
computation of the number of microorganisms per volume was recorded.
Differences
in the findings between the “swish” analysis compared to the “irrigation and
swish” compared to the pocket micropipette analysis present conflicting
results. The direct pocket analysis provides the best means of determining
which bacteria predominate and/or co-exist in healthy and diseased patients’
periodontal tissues. The predominance of type and category of bacteria and the
changes from health to varying stages of disease are presented.
There is
a shift from a more aerobic and facultative anaerobic Gram-positive biofilm in
healthy pockets that are replaced by anaerobic and Gram-negative biofilm found
in periodontal disease. The difference starts at the 3 mm pocket depth between
patients without periodontal disease versus patients with periodontal disease.
Treatment results can be appraised by comparing the microbiome components to
that found in health as compared to components found in disease. This is a
small sample and additional investigations may be needed to confirm the
findings of this study.
Keywords: Microorganisms, Periodontal, Microbiome, Gram-negative
INTRODUCTION
Biofilms
related to periodontal disease have been evaluated by various means. Research
using a checkerboard DNA-DNA hybridization to identify 40 different bacterial
strains shows the predominant initial colonizers of the oral environment are
Gram-positive facultative anaerobic cocci and rods, including Streptococcus and
Actinomyces species and aerobic bacteria [1]. These initial colonizers provide
a foundation for further development of dental biofilm. Early microbial
succession involved mainly Gram-positive and Gram-negative aerobic and
facultative anaerobes with a few Gram-positive anaerobes [2].
These findings indicate that typical
co-colonization of specific oral species,
among which a cluster with the nomenclature “red
complex” composed of the Gram-negative anaerobic species Tannerella forsythia, P.
gingivalis and Treponema denticola,
are associated with increased pocket depth and bleeding upon clinical pocket
probing, Socransky et al. [5] report the other four clusters examined were not
shown to be associated with clinical parameters indicating periodontal disease.
Other
possible explanations about the shift from health to periodontitis involves
when a low number of bacteria (102-103) that are mostly
Gram-positive aerobic and facultative anaerobes increase in number and are
overtaken by a greater number (104-105) of Gram-negative
anaerobic microorganisms [6]. Gram-positive and Gram-negative bacteria equally
induce IL-1 beta, but Gram-positive bacteria generate twice as much TNF-alpha.
Gram-negative bacteria induce at least twice as much IL-6 and IL-8 [7]. The
increased incidence of Gram-negative anaerobic bacteria induces systemic
challenges and immune system responses.
Gram-negative
bacteria produce lipopolysaccharides (LPS) that induce inflammatory cell
infiltrate in the blood vessel walls, causing vascular smooth muscle
proliferation, vascular fatty degeneration and intravascular coagulations. LPS
up-regulates endothelial cell adhesion molecule expression and increases the
secretion of interleukin-1 (IL-1), tumor necrosis factor alpha (TNF-alpha) and
thromboxane, which increases platelet aggregation and adhesion, causing the
formation of lipid laden foam cells and deposits of cholesterol and cholesterol
esters [8].
The severity
of the host response to both Gram-negative and Gram-positive bacteria plays a
major role in causing inflammation and tissue sepsis. These bacteria produce a
range of virulence factors that enable them to escape the immune defenses and
disseminate to remote organs. Toxins that interact with host cells via specific
receptors on the cell surface trigger a dysregulated immune response [9]. Gram-negative
bacteria pose more host inflammatory complications due to:
1.
There
is a membrane present around the cell wall of gram-negative bacteria which
increases the risk of toxicity to the host, but this membrane is absent in
gram-positive bacteria.
2.
Porin
channels are present in gram-negative bacteria which can prevent the entry of
harmful chemicals and antibiotics like penicillin. These channels can also
expel out antibiotics making it more difficult to treat in comparison to
gram-positive bacteria.
3.
The
risk of resistance against antibiotics is higher in Gram-negative bacteria due
to the presence of external covering around the cell wall.
4.
Gram-negative
bacteria possess both exotoxins and endotoxins but in case of gram-positive
bacteria there are individual exotoxins [10].
Another
possible explanation of the shift in the biofilm relates to oxygen levels in
the periodontal pockets. A relationship exists between the subgingival microbes
and the oxygen tension in periodontal pockets, suggesting anaerobes increase as
the pocket depth increases and oxygen tensions decrease [11]. Anaerobic
bacteria are shown to be resistant to short-term periodontal therapy [12] and
can regrow in a matter of days, continuing the infectious process [13].
Critical
wound colonization is a term utilized to express wound chronicity as it relates
to the quantity and quality of the infectious agents as well as the host
responses [14]. An increased number of more virulent bacteria commonly relates
to an increase in the host inflammatory responses. This involves a framework
where virulent immune provoking behaviors and enhanced immune resistance enables
invading pathogens to overcome resident microorganisms [15]. Pathogenic
bacteria succeed by creating a novel immunologic challenge to which they are
already adapted. Decreasing the number of pathogens reduces virulence, while
specific bacteria are associated with higher virulence [16].
Oral saliva
diagnostics provide information on a variety of pathogens, but many of these
systems are limited in scope and whether the bacteria originate from the
periodontal tissues or other oral structures is questioned. There are no
FDA-approved salivary diagnostic tests for evaluating the risk of periodontal
disease [17]. Site specific diagnostics can determine the pathogens present in
the periodontal infection and this more precise information may be essential to
customize the proper corrective measures and determine treatment results.
This article
discusses the biofilm constituents that appear from oral saliva analysis
compared to the biofilm present in the periodontal pocket by micropipette
analysis. There is a discrepancy between the findings between a saliva analysis
and a micropipette analysis. These differences may have a bearing on treatment
options and determining treatment success or failure.
METHOD
Three
methods of biofilm analysis were compared in this study. All evaluators were
calibrated for reliability and accuracy. Oral saliva analysis, lavage and oral
saliva analysis and direct pocket pathogen analysis by micropipette collection
are examined by DNA analysis to determine bacterial presence in healthy and diseased
periodontal pockets. The results of these different analysis systems were also
compared. This pilot study involves 73 periodontal sulcus samples collected by
micro-pipette, 14 (lavage and swish) and 16 (swish only) samples submitted for
DNA analysis (MicroGenDx) to evaluate the composition and characteristics of
the biofilm. Only the bacteria and fungi that comprise 2% or more of the periodontal
biofilm were identified. Partial percentages are rounded off to the closest
whole number.
Patients were divided into those without
periodontal disease (3 mm pockets or less with no BOP) and those with
periodontal disease (3 mm or more with bleeding). The DNA analysis provided the
percentage and number of Gram-positive versus Gram-negative bacteria and
whether the bacteria were anaerobic, facultative anaerobes or aerobic. Fungi
were also reported.
The compositions of biofilms in healthy
patients were evaluated by micropipette analysis to determine what bacteria comprise
the biofilm in healthy pockets. These findings were compared to the biofilm
found in diseased patient’s pockets to determine possible patterns of
pathogenicity. The biofilm determined by micropipette collection were compared
to oral saliva analysis. Two means of saliva analysis were utilized; “swish”
saliva samples of the entire mouth and a second irrigation of the periodontal
pockets, swish and saliva collection to evaluate the biofilm.
A swish only saliva collection and a lavage
and swish saliva collection were completed and all bacteria and fungus 2% or
more were computed as well as the number of bacteria/volumes. Samples that are
found 1% of the time or less are not compared, so results will often
demonstrate less than 100% in the evaluation.
In the “swish” sample, 5 cc of sterile saline
was swished in the patient’s mouth for 60 seconds and then collected for
analysis in a sterile container. In the second salivary analysis, 5 cc of
sterile saline was placed into an irrigation syringe which was used to lavage
the periodontal pockets. This irrigation-swished material was swished and
maintained in the patient’s mouth for 60 s and then expectorated into a sterile
container for shipment for biofilm analysis.
Individual periodontal pockets were analyzed
by micro-pipette suction to remove the biofilm from periodontal sulcus or
pocket. A blunt tipped needle attached to a syringe was inserted to the depth
of the periodontal pocket and the syringe plunger was slightly elevated for 10
seconds to create a negative pressure within the syringe, so the microbiome was
“sucked” into the tip. The tip was removed from the pocket with the plunger
elevated so all the sample remained in the tip. The micro-pipette tip was
removed from the syringe and placed in a sterile labeled transport container
for shipment to MicroGenDx for analysis of the biofilm.
MicroGenDx evaluated all samples as to the
composition of the biofilm. All samples were found to contain adequate biofilm
for analysis. MicroGenDx recorded the concentration of the biofilm as low (10X3-5),
medium (10 X5-7) and high (10 X7>). The numeric
compositions used to determine the bacteria/volume were computed as 10X3,
10X5 and 10X7. All microorganisms that comprise 2% of the
population or greater were recorded. Comparisons were made between healthy
pockets (pocket probing depths 3 mm or less without BOP) that were compared to
patients clinically determined to have periodontal disease.
Multiple bacteria were found in this study
and divided into distinct groupings: Gram positive or Gram negative. The
samples were also categorized as fungi or types of bacteria: anaerobes,
facultative anaerobes and aerobic bacteria to the genus/species level.
Subspecies were not determined. The number of bacteria was also evaluated to
determine the population density per volume according to the sample technique.
The oral solutions “swish and expectorate”
was gathered first. Second the oral rinse solution was irrigated into the
deeper periodontal pockets and swished and collected. Micro-pipette samples of
periodontal pockets were gathered after the saliva samples were collected. All
samples were labeled and shipped for analysis.
All samples were found to contain a minimum
of 10X3 or greater. The type of bacteria and fungi were evaluated
for different pocket depths and compared to the oral saliva sample techniques.
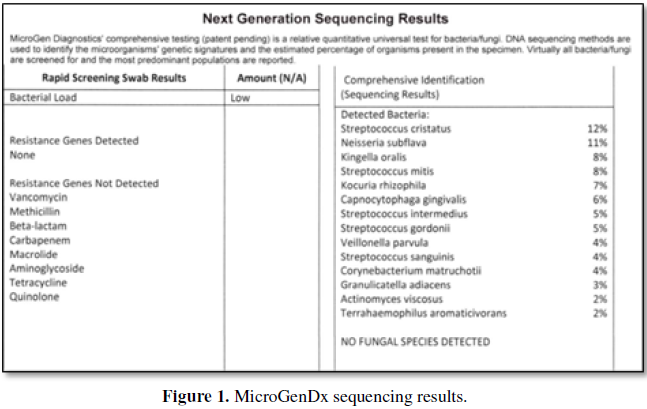
The following is an example of a MicroGenDx report (Figure 1).
The percent number of the individual type
(genus/species) was first categorized as Gram positive versus Gram negative.
The sample was further analyzed by the percentage of anaerobes, facultative
anaerobes and aerobic bacteria. Bacteria that were less than 2% of the
population were not considered in the evaluations. In the above sample no fungi
were discovered.
The percentage presence of Gram positive
bacteria in the above example are the Streptococcus, Kocuria, Granulicartella
and Actinomyces, comprising 42% of the population. Gram negative species
(remaining) comprise 39% of the population with the remaining 19% existing in
less than 2% concentrations. Anaerobes, facultative anaerobes and aerobic
percentages of the total population would be: 19%, 41% and 21%. No fungi were
present in this sample. The percentages of the specific categories may be
depicted in graphic representation (Figure
2).
Yellow
Gram
positive
Pink Gram
negative
Red Anaerobes
Green Facultative
anaerobes
Blue Aerobes
Black Fungi
The graphic representation of the MicroGenDx
sample above is illustrated in Figure 2.
RESULTS
The results of this study help determine the
microbes found in healthy tissues and evaluate changes in the periodontal
microbiome as periodontal pocket depth and pathology increase. The initial
evaluations are completed for patients without periodontal disease (PD) and
where no pockets are greater than 3 mm. The initial tests involved an oral
“swish” (Figure 3), “irrigation and
swish” (Figure 4) and a micropipette
analysis (Figure 5) of the
microbiome.
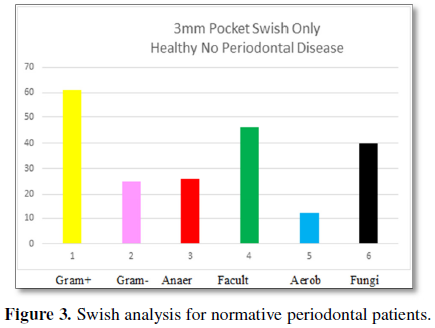
The “swish” samples of 3 mm pockets for
patients free of periodontal disease are presented in Figure 3. The response to a Gram stain divides the sample into two
types of microorganisms, those which are Gram-positive and those which are
Gram-negative. Only those bacteria present 2% of the time are greater are
included in this and all following analyzes.
Gram-positive bacteria are present at 61% and
Gram-negative are present at 25%. The categories of bacteria found are
anaerobes at 26%, facultative anaerobes as the predominant species at 46% and
aerobic bacteria at 12%. Fungi are found in 40% of the patient’s “swish”
samples. The number of bacteria/volume is 10 × 5.8. These findings can be
compared to the “irrigation and swish” samples for periodontal healthy patients
3 mm pockets which are presented in Figure
4.
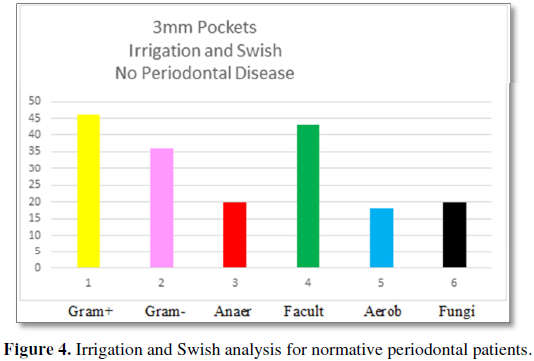
The “irrigation and swish” samples for
healthy patients demonstrate Gram-positive bacteria are present 46% compared to
36% for Gram-negative bacteria. The categories of bacteria are 20% anaerobic,
43% facultative anaerobes and 18% aerobic bacteria with a 20% incidence of
fungi. The number of bacteria/volume is 10 × 5.5. These results are compared to
the bacteria found in the healthy periodontal sulci by micropipette analysis.
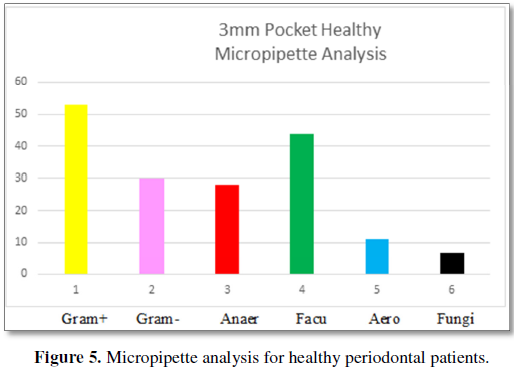
Figure 5 demonstrates the bacteria found
in 3 mm periodontal pockets for patients without periodontal disease. Gram-
positive bacteria are present at 53% and are more prevalent than Gram-negative
bacteria at 30%. There are more facultative anaerobes 44% than anaerobes 28%,
with aerobic bacteria present at 11%. Fungi are present at 7%. The number of
bacteria is 10 × 4.0.
Table 1 compares the microorganisms found
in the saliva “swish” sample, “irrigation and swish” sample and the
micropipette analysis samples for patients without periodontal disease. The
predominant types of bacteria in all samples are Gram-positive bacteria. All
three samples demonstrate the predominant category of bacteria is facultative
anaerobes, followed by anaerobic bacteria, and aerobic bacteria. Fungi are
evident in the both oral saliva samples and in the micropipette analysis at
varying proportions. There is more bacteria/area in the saliva samples as
compared to the micropipette analysis. The predominant species in healthy
periodontal tissues and in the oral saliva samples of healthy periodontal
patients are Gram-positive and facultative anaerobes with a lesser percent
presence of anaerobes and aerobic bacteria and some fungi.
Three-millimeter
pockets are accepted as normal regarding periodontal health [18]. Patients with
periodontal disease somewhere in their mouth also have 3 mm pockets. The
composite of the biofilm of patients 3 mm pockets with periodontal disease are
analyzed by “swish”, “irrigation and swish” and micropipette analysis of 3 mm
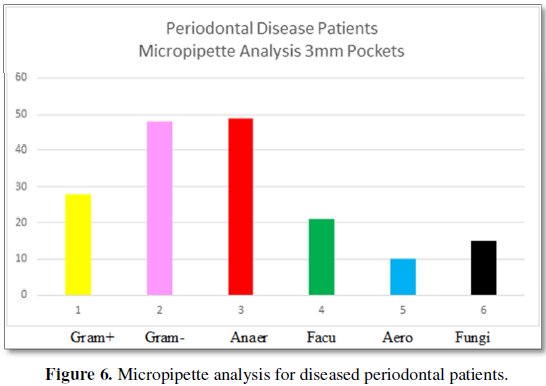
pockets for patients with periodontal disease are presented in Figure 6.
Figure 6 graphically represents bacteria
presence in 3 mm periodontal pockets from patients diagnosed with periodontal
disease somewhere in their mouth by micropipette analysis. Gram positive
microbes are present at 27% as compared to Gram-negative present at 50%.
Anaerobes constitute 48% of the population, facultative anaerobes comprise 20%
and aerobic bacteria are present 11% of the time. Fungi are found at 15%. The
number of bacteria is 10 × 3.6.
There are distinct differences between the
microbiome in healthy 3 mm periodontal pockets and 3 mm periodontal pockets
from patients with periodontal disease. These differences are presented in Table 2. Table 2 compares the bacteria present in 3 mm pockets of patients
free of periodontal disease to the bacteria found in 3 mm pockets of patients
who have periodontal disease somewhere in their mouth.
Table 2 demonstrates differences in the microbiome
by micropipette analysis found in 3 mm pockets of patients with and without
periodontal disease. Gram-positive bacteria predominate in healthy 3 mm
pockets, while Gram-negative bacteria predominate in 3 mm pockets of
periodontal disease patients. Facultative anaerobes are the predominant species
in healthy 3 mm periodontal pockets, while anaerobes predominate in 3 mm
periodontal pockets of patients with periodontal disease. Aerobic bacteria
remain constant for both groups and a small percentage of fungi are found in
health 3 mm pockets and in 3 mm pockets of periodontal disease patients.
Periodontal pockets greater than 3 mm are
determined in this study to be evidence of periodontal disease. These are
evaluated by oral saliva “swish”, “irrigation and swish” samples and
micropipette analysis. Oral saliva samples “swish” and “irrigation and swish”
are taken for all patients found to have periodontal disease. The composite of
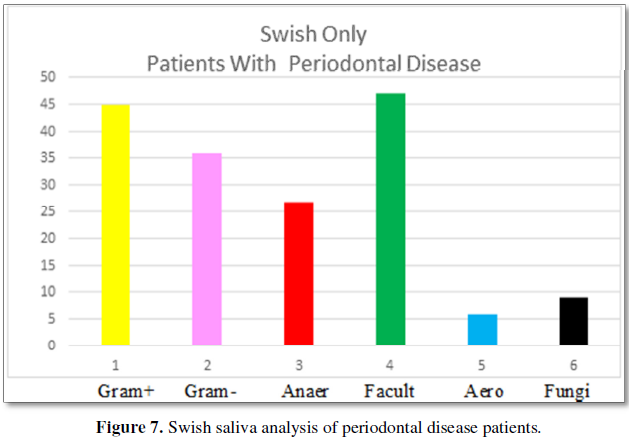
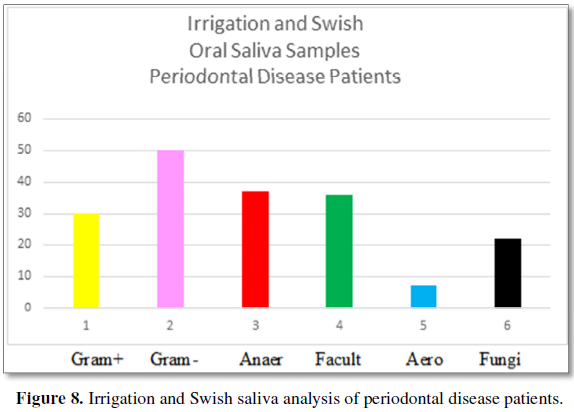
the oral saliva findings are presented in Figure
7 “swish” and Figure 8
“irrigation and swith” sample results.
Figure 7 demonstrates the composition of the biofilm found in
oral saliva “swish” analyzes for patients with periodontal disease.
Gram-positive bacteria are evident at 45% and are more prevalent than
Gram-negative bacteria at 36%. Facultative anaerobes predominate at 47% with
anaerobes at 27% and aerobic bacteria at 6%. Fungi are present at 9%. The
number of bacteria per volume is 10 × 6.6.
Figure 8 demonstrates the biofilm
composition of oral saliva samples using the “irrigation and swish” technique.
Gram-positive bacteria are found at 30% and Gram-negative bacteria are found at
50%. 20% of the bacteria are present in concentrations of less than 2%.
Anaerobes are found at 37% with facultative anaerobes at 36% and aerobic
bacteria at 7%. Fungi are present at 22%. The number of bacteria per volume is
10 × 6.8.
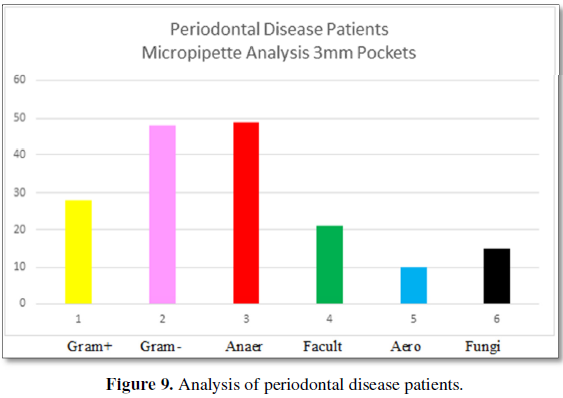
Figure 9 demonstrates the bacteria found
in 3 mm pockets of patients with periodontal disease somewhere in their mouth.
The types of bacteria are 27% Gram-positive and 50% are Gram-negative. The
categories of bacteria are 48% are anaerobic, 20% are facultative anaerobes and
11% are aerobic bacteria. Fungi are present at 15%. The number of
bacteria/volume is 10 × 3.6.
Differences are evident in these findings. The oral “swish” analysis demonstrates Gram-positive and facultative anaerobes are the predominant species in patients with periodontal disease. The “irrigation and swish” analysis demonstrates Gram-negative bacteria and an almost equal number of facultative anaerobes and anaerobes predominate. The micropipette analysis of 3 mm periodontal pockets from patients with periodontal disease demonstrates Gram-negative and anaerobic bacteria predominate.
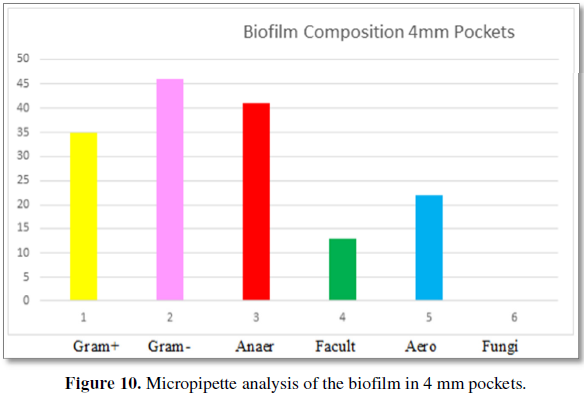
Figure 10 demonstrates the bacteria found by
micropipette analysis from patients 4 mm periodontal pockets. The type of
bacteria are 35% Gram-positive and 46% Gram-negative. The categories of
bacteria are 41% are anaerobic, 13% are facultative anaerobes and 22% are
aerobic bacteria. No fungi are found in 4 mm pockets. The number of bacteria
per volume is 10 × 4.3.
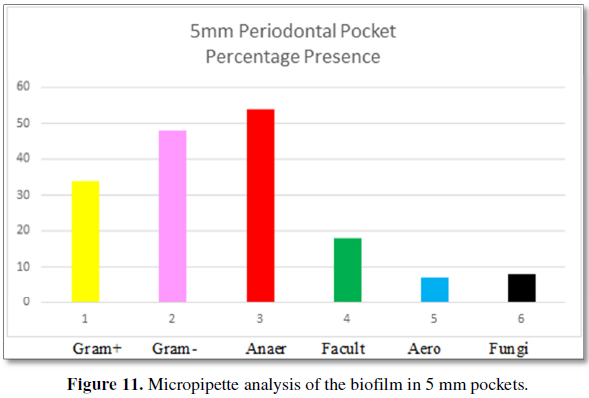
Figure 11 demonstrates the bacteria found by
micropipette analysis from patient’s 5 mm periodontal pockets. The type of
bacteria are 34% Gram-positive and 48% Gram-negative. The categories of
bacteria are: 54% are anaerobic, 18% are facultative anaerobes and 7% are
aerobic bacteria. Fungi are found at 8% in 5 mm pockets. The number of bacteria
per volume is 10 × 4.8.
Figure 12 demonstrates the bacteria found by micropipette
analysis from patients 6 mm periodontal pockets. The types of bacteria are: 10%
Gram-positive and 68% Gram-negative. The categories of bacteria are: 64% are
anaerobic, 9% are facultative anaerobes and 4% are aerobic bacteria. Fungi are
absent from 6 mm pockets. The number of bacteria per volume is 10 × 4.7.
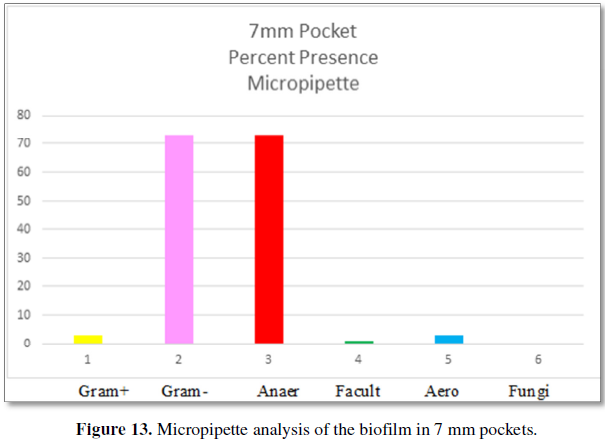
Figure 13 demonstrates the bacteria found by
micropipette analysis from patients 7 mm periodontal pockets. The types of
bacteria are: 3% Gram-positive and 73% Gram-negative. These are the bacteria
that comprise at least 2% of the total biofilm consistency. The categories of
bacteria are: 73% are anaerobic, 1% is facultative anaerobes and 3% are aerobic
bacteria. Fungi are absent from 7 mm pockets. The number of bacteria per volume
is 10 × 4.6.
Comparison of the type of bacteria and category
of bacteria for the oral saliva analyzes “swish”, “irrigation and swish” and
the microbiome found by micropipette analyzes for all periodontal pockets from
periodontal disease patients are presented in Figures 14A and 14B.
Gram-positive
bacteria are the predominant species in the oral “swish” method of evaluation.
Gram-negative bacteria are the predominant species in the oral “irrigation and
swish” method of evaluation. The micropipette analysis demonstrates
Gram-negative bacteria are the predominant species in the 4-7 mm pockets and
the incidence increases as the pocket depth increases.
Figure 14B demonstrates that facultative
anaerobes are the predominant species in the oral “swish” analysis. The
predominant species in the “irrigation and swish” are facultative anaerobes and
anaerobic bacteria. The predominant category of microbes in 4-7 mm pockets are
anaerobic bacteria that increase in incidence as the pocket depth increases.
Figures 14 A and 14B illustrates the type and category
of bacteria present in each sample for patients with periodontal disease. The
oral saliva “swish” sample, “irrigation and swish” and the micropipette
constituents for periodontal pockets of 4, 5, 6 and 7 mm are compared to
evaluate similarities or differences. There is a significant difference in the
“swish” analysis as the predominant species are Gram-positive bacteria, where
the “irrigation and swish” and the micropipette analyzes demonstrate the
predominant species are Gram-negative. The micropipette results for 4-7 mm
pockets demonstrate the increased predominance of Gram-negative bacteria and
the decrease in Gram-positive bacteria as the pocket depth increases.
Figure 14B demonstrates the “swish” analysis predominant category of bacteria is facultative anaerobes, where the “irrigation and swish” predominate category is facultative anaerobes and anaerobic bacteria. The micropipette analysis of 4-7 mm pockets predominant category demonstrates a steady increase in the anaerobic bacteria as the facultative anaerobic and aerobic bacteria decrease. The discrepancy between the findings between the “swish” analysis, “irrigation and swish” and the micropipette analyzes raise concerns as the different analyzes demonstrate difference predominant species and types of bacteria but were gathered from the same patients.
DISCUSSION
Knowing what comprises the biofilm in healthy
periodontal tissues is important. The healthy biofilm serves as a treatment
goal. Understanding the changes that occur in disease helps clarify disease
etiology. The evaluation means to make these determinations must be consistent
and accurate. There is a similarity of diagnostic results between oral saliva
analyzes “swish” irrigation and “swish” and micropipette analysis when
evaluating healthy periodontal tissues. The analyses find a predominance of
Gram-positive and facultative anaerobes in healthy conditions.
The similarities diverge when evaluating periodontal
disease tissues with the “swish”, “irrigation and swish” and a micropipette
analysis. The predominant species in periodontal disease oral saliva “swish”
analysis are Gram-positive and facultative anaerobes. The predominant species
in the oral saliva “irrigation and swish” analysis are Gram-negative anaerobic
and facultative anaerobes. The predominant species in the micropipette analysis
of 4-7 mm pockets shows an increasing incidence of Gram-negative and anaerobic
bacteria. The difference between the three analysis techniques raises a
question of accuracy and reliability.
One reason for the differences is the area
evaluated. The “swish” analysis samples the entire oral area, resulting in
bacteria from periodontal tissues, but also bacteria that are found on all
other oral structures. The “irrigation and swish” sampling lavages the
periodontal pocket, but also collects the bacteria from all other oral
structures. The micropipette analysis only evaluates the microbiome of the
periodontal pocket. The consistency of the micropipette results supports this
method as the most accurate representation of the biofilm in the periodontal
pocket.
The differences in predominance of the type
and category of bacteria in the micropipette analysis begins in 3 mm pockets
for patients with periodontal disease as compared to 3 mm pockets of patients
without periodontal disease. The micropipette microbiome in the diseased pocket
demonstrates an increasing incidence of Gram-negative and anaerobic bacteria as
the pocket depth increases.
Evaluation of treatment success may be
misleading with the differences of the three methods. When evaluating
periodontal disease patients, the “swish” analysis demonstrates a predominance
of Gram-positive and facultative anaerobe while the “irrigation and swish”
technique demonstrates a predominance of Gram-negative and equal anaerobic and
facultative anaerobes. The micropipette analysis of the periodontal disease
pockets demonstrates a predominance of Gram-negative and anaerobic bacteria in
diseased pockets that increases as the pocket depth increases. Different
evaluation methods of the same patients should coincide, not diverge. If the
micropipette analysis is the most accurate, the “irrigation and swish” is the
next most accurate and the most inaccurate is the “swish” analysis. This study
is a small sample and these results should be evaluated in larger studies with
a greater in-depth analysis.
CONCLUSION
This research helps clarify the biofilm found
in healthy periodontal tissues, which varies significantly from the biofilm
found in diseased tissues. It is important to know what biofilm constituents
exist in the host tissues to determine health or disease as this may be
important in determining treatment success or failure. The three methods of
evaluating the biofilm, “swish”, “irrigation and swish” and micropipette
analysis coincide with regard to the biofilm in healthy tissues, but the
results vary and are uncertain when evaluating the etiology of disease.
Three methods of analysis are compared in
this study and all three methods (“swish”, “irrigation and swish” and
micropipette) demonstrate similar findings with healthy periodontal tissue. The
three analyzes for healthy periodontal tissues (3 mm or less with no BOP)
demonstrates Gram-positive bacteria and facultative anaerobes predominate.
There are lesser amounts of anaerobic bacteria, aerobic bacteria and fungi. All
three analysis methods generally agree with regard to healthy tissues, but this
similarity is missing with regard to diseased tissues.
The three methods evaluate the type of
bacteria; Gram-positive or Gram-negative. The “swish” analysis for patients
with periodontal disease demonstrates a predominance of Gram-positive bacteria.
The “irrigation and swish” analysis of patients with periodontal disease and
the micropipette analysis of periodontal pockets 4 mm or greater demonstrate a
predominance of Gram-negative bacteria. The micropipette analysis demonstrates
Gram-negative bacterial predominance increases as pocket depth increases.
The categories of bacteria vary in the
analysis of periodontal disease patients between the “swish”, “irrigation and
swish” and the micropipette techniques. The “swish” analysis of periodontal
disease patients demonstrates facultative anaerobes predominate, followed by
anaerobes, aerobic bacteria and fungi. The “irrigation and swish” analysis of
periodontal disease patients demonstrates a comparable facultative and
anaerobic population with a lesser presence of aerobic bacteria and fungi. The
micropipette analysis demonstrates anaerobic bacteria predominate at 3 mm in
periodontal disease patients and the predominance increases as the pocket depth
increases.
The variability of the findings is troubling since the tests were completed on the same patients. One would expect the findings to coincide, but this is not what occurred. Comparison of the oral saliva “swish” test with the “irrigation and swish” and the micropipette analysis of the type and category of bacteria demonstrates significant imprecisions. If the micropipette analysis best represents the microbiome in the periodontal pocket, the “irrigation and swish” analysis is less accurate and the “swish” analysis provides the most inaccurate information.
1. Li J, Helmerhorst EJ, Leone CW, Troxler
RF, Yaskell T, et al. (2004) Identification of early microbial colonizers in
human dental biofilm. J Appl Micobiol 97: 1311-1318.
2. Teles FR, Teles RP, Uzel NG, Song
XQ, Torresyap G, et al. (2012) Early microbial succession in re-developing
dental biofilms in periodontal health and disease. J Periodontol Res 47:
95-104.
3. Berezow AB, Darveau RP (2000)
Microbial shift and periodontitis. Periodontol 55: 36-47.
4. Uematsu H, Hoshino E (1992)
Predominant obligate anaerobes in human periodontal pockets. J Periodontal Res
27: 15-19.
5. Socransky SS, Haffajee AD, Cugini
M, Smith C, Kent RL Jr. (1998) Microbial complexes in subgingival plaque. J
Clin Periodontol 25: 134-144.
6. Alwaeli AZJ (2018) Anaerobic
bacteria associated with periodontitis. Available at: https://www.intechopen.com/books/oral-microbiology-in-periodontitis/anaerobic-bacteria-associated-with-periodontitis
7. Hessle CC, Andersson B, Wold AE
(2005) Gram-positive and gram-negative bacteria elicit different patterns of
pro-inflammatory cytokines in human monocytes. Cytokine 30: 311-318.
8. Dahiya S, Kaur T, Srivastava A
(2018) Oral foci of infection leading to systemic diseases - An emerging
problem in medicine. Eur J Pharm Med Res 5: 167-171.
9. Ramachandran G (2014)
Gram-positive and gram-negative bacteria toxins in sepsis. Virulence 5:
213-218.
10. Hayat K (2013) Why is it more
difficult to treat gram negative bacteria. Available at: https://medimoon.com/04/why-is-it-more-difficult-to-treat-gram-negative-bacteria/
11. Loesche, WJ, Gusberti F Mettraux
G, Higgins T, Syed S (1983) Relationship between oxygen tension and subgingival
bacteria flora in untreated human periodontal pockets. Infect Immun 42:
659-667.
12. Camargo GA, Abreu MG, Cordeiro RS,
Wenderoscky Lde F, Duque C (2016) Prevalence of periodontopathogens and Candida spp. in smokers after non-surgical
periodontal therapy - A pilot study. Bras Oral Res 30: e92.
13. Teles FR, Teles RP, Uzel NG, Song
XQ, Torresyap G, et al. (2012) Early microbial succession in re-developing
dental biofilms in periodontal health and disease. J Periodontal Res 47: 95-104.
14. White R, Cutting K (2008) Critical
colonization of chronic wounds: Microbial mechanisms. Wounds UK 4: 70-78.
15. Brown SP, Le Chat L, Taddei F
(2008) Evaluation of virulence; triggering host inflammation allows invading
pathogens to exclude competitors. Ecol Lett 11: 44-51.
16. Gandon S, Michalakis Y (2000)
Evolution of parasitic virulence against qualitative or quantitative host
resistance. Proc Biol Sci 267: 985-990.
17. https://www.ada.org/en/member-center/oral-health-topics/salivary-diagnostics
QUICK LINKS
- SUBMIT MANUSCRIPT
- RECOMMEND THE JOURNAL
-
SUBSCRIBE FOR ALERTS
RELATED JOURNALS
- Journal of Carcinogenesis and Mutagenesis Research (ISSN: 2643-0541)
- International Journal of Diabetes (ISSN: 2644-3031)
- Journal of Neurosurgery Imaging and Techniques (ISSN:2473-1943)
- Journal of Cancer Science and Treatment (ISSN:2641-7472)
- Archive of Obstetrics Gynecology and Reproductive Medicine (ISSN:2640-2297)
- Journal of Ageing and Restorative Medicine (ISSN:2637-7403)
- Chemotherapy Research Journal (ISSN:2642-0236)