904
Views & Citations10
Likes & Shares
INTRODUCTION
The US National Cancer Institute (NCI) and the National Human Genome Research Institute (NHGRI) created the Cancer Genome Atlas (TCGA) Project in 2006 with bulk tumor of cellular heterogeneity with one-time point [1]. However, it is essential to track down subclonal evolution of cancer stem cells (CSCs) evolving with treatment, such as Temodar®-driven mutations [2,3]. Indeed, Darwinian pressures arising from systemic therapy, result in the clonal selection of initially rare subclone variants within a tumor. Novel technologies such as single cell circulating tumor cell sequencing would allow real-time monitoring of clonal heterogeneity, including Loss of heterozygosity (LOH) assays at genetic DNA level, at primary and metastatic sites, as well as for improved targeted early intervention, better care, prognosis, tumor subclonal recognition with immunotherapy [4]. This approach is of particular interest in highly lethal cancers like glioblastomas and colorectal adenocarcinomas, which express high intratumoral heterogeneity when a single signaling network is observed [5,6]. Therefore, advances in transcriptome analysis at single-cell resolutions have become increasingly sought out tools to investigate causative signal pathways alterations in cancer, as they can specifically address the shortcomings of studying bulk lysates for heterogeneous tumor biopsies [7]. Single cell stem cells were very tough to extract from heterogeneous populations of tumour, however; recent innovative single-cell sequencing to profile the gene-expression landscapes of more than 20,000 cells in the motor cortex of brain [8], makes it possible for complex tumors to track the subclonal evolution through phylogenetic analysis [9].
First, it is essential to define the Head and Neck Squamous Cell Cancer (HNSCC) stem cells by the biomarkers. The aldehyde dehydrogenase (ALDH) is a polymorphic enzyme responsible for the oxidation of aldehydes to carboxylic acids. ALDH can be used in the identification of CSCs [16] in HNSCC. It has been shown that stem cells express elevated levels of ALDH. Chemoresistant cancer cells express high levels of ALDHs, particularly in HNSCC. The ALDH family of enzymes detoxifies both exogenous and endogenous aldehydes. Since many chemotherapeutic agents, such as cisplatin, result in the generation of cytotoxic aldehydes and oxidative stress, we hypothesized that cells expressing elevated levels of ALDH may be more chemoresistant due to their increased detoxifying capacity and that inhibitors of ALDH may sensitize them to these drugs. In another word, HNSCC-CSCs can be identified by high cell-surface expression of CD44 and high intracellular activity of ALDH, termed ALDHhighCD44high [17]. Microarray analysis of cisplatin-resistant ALDHhighCD44high cells indicates that their signaling pathways have significant implications for the pathobiology of cancer (e.g. TNFα, IFN, IL6/STAT, NF-κB, FGF2) [17]. The activation of ALDH3A1 by a small molecule activator (Alda-89) increased survival of ALDHhigh HNSCC cells treated with cisplatin while treatment with a novel small molecule ALDH inhibitor (Aldi-6) resulted in a marked decrease in cell viability, suggesting a promising strategy [18].
Second, HNSCC cells that are resistant to chemotherapy, lead to tumor recurrent or metastatic that has the poor prognosis with less than 1 year median survival and [17]. Autologous CSCs for the screening of personalized treatment (precision medicine) can be derived from HNSCC pateint specimens. Studies in our lab have revealed that both CSC and its related tumor microenvironment can be used for therapeutic detection before administration to patients. We think that tumor surveillance and response by these patient’s autologous CSC screening help determine how patient’s CSC react and evolve with therapeutics, i.e., co-evolution of CSCs with therapeutics. Reports from brain tumor stem cells in glioma patients [7] and in primitive neuroectodermal tumor [19,20] with single-cell transcriptome for relapse prognosis [21] and in situ hybridization [22] prompted us to realize that patient’s HNSCC CSCs (HNCSC) possess a capacity for tumor evolution of therapeutics as such development shows prognostic value for patients. With upregulation of vascular endothelial growth factor (VEGF) and VEGF receptor 2 (VEGFR2) of highly angiogenic phenotypes, tumor endothelial cells (TECs) exhibit higher proliferative and migratory capacity, compared with those of normal endothelial cells (NECs). Such TECs show ALDHhigh cell populations by fluorescence-activated cell sorting (FACS) [23]. Sheng et al. [24] demonstrated that with decreased ALDH activity, the expression levels of stemness-associated markers, CD133+, octamer-binding transcription facto 4 (Oct4) and sex determining region Y box 2 also reduced. They also showed, “an increased number of mice developed tumors in the ALDHhigh group 16 weeks following the injection of 500 cells, whereas tumors appeared at eight weeks in the ALDHlow group”. The mice in the ALDHneg group exhibited less tumor formation under these conditions.” They conclude that “ALDHhigh cells had characteristics of self-renewal ability, in a relative resting stage; while the ALDHlow cells had characteristics of GCPCs with limited self-renewal ability, but were in a rapid proliferation stage” [24]. Thus, HNCSC cells can be identified with drugs sensitive to ALDH(high)+ cells and isolated with their resistance to fluorouracil (5-FU) in vitro and in vivo, while tumor endothelial cells (TEC) can be identified with high ALDH activity (ALDHhigh), along with upregulation of stem-related genes such as multidrug resistance 1, CD90, ALP, Oct-4, Platelet-derived growth factor (PDGF)-A [25].
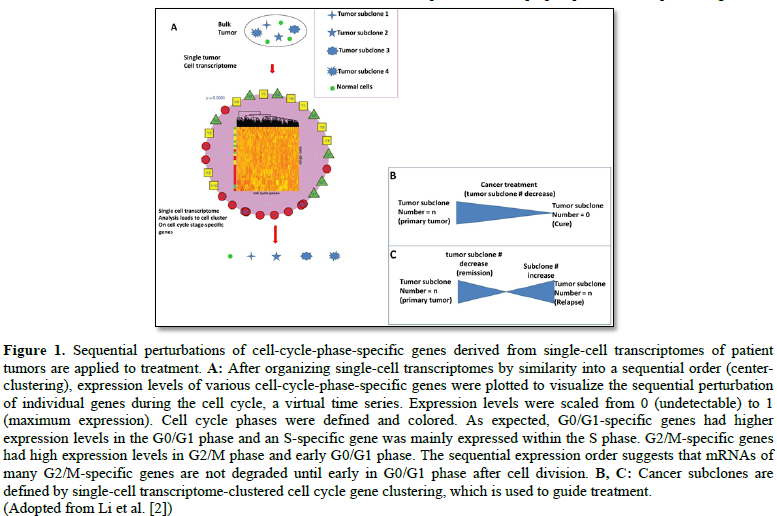
Third, as well documented in glioblastomas [26], medulloblastoma [27], leukemia [28] and germ cell tumors [29]: all of these cancer types evolve with treatment (radiation and chemotherapy) surviving by coming up with new mutations in subclonal evolution, which can be defined by single-cell transcriptome technology (Figure 1). For example, “Although well-tolerated, the efficacy of bevacizumab was somewhat disappointing, possibly due to the high rate of secondary high-grade gliomas in the studied patient cohort and the late use of bevacizumab in the course of the disease” [30]. Following analysis of tumor specimens, distinct molecular pathogenesis of secondary tumor arising after radiation therapy was determined by cancer genome-scale technology for genomic mutation signatures, particularly, discovered in secondary neoplasms after cranial or craniospinal radiation in high-grade astrocytomas that prognoses for poor clinical outcomes [31]. Surprisingly, this cohort had “a high frequency of TP53 mutations, CDK4 amplification or CDKN2A homozygous deletion and amplification or rearrangements involving receptor tyrosine kinase and Ras-Raf-MAP kinase pathway genes including PDGFRA, MET, BRAF and RRAS2,” but, “lacked alterations in IDH1, IDH2, H3F3A, HIST1H3B, HIST1H3C, TERT (including promoter region) and PTEN, which genetically define the major subtypes of diffuse gliomas in children and adults.” Such subclonal changes can be tracked down using single-cell RNA sequencing, as shown in 3321 single-cells from six primary H3K27M-glioma and matched models, for the discovery of “oncogenic and developmental programs in H3K27M-glioma at single-cell resolution and across genetic subclones.” This subclonal tracking surfaces a therapeutic window [32] on potential therapeutic targets.
To identify a therapeutic window [32] on potential therapeutic targets, we need to define spatiotemporal expression patterns of new biomarkers from HNSCC to significantly improve the efficacy of therapies. As such a new biomarker, known a molecular mechanism, AF4/FMR2 family member 4 (AFF4), the core component of Super elongation complex (SEC), is upregulated dramatically in HNSCC, which is a potential target of therapies for patients with HNSCC [33]. Besides, Disulfiram (DS) has been reported as an inhibitor of ALDH and increasing studies showed it has anti-cancer effects in a copper (Cu)-dependent manner [34]. As “DS/Cu inhibited the expression of stem cell transcription factors NANOG and OCT4, and abolished the clonogenicity of multiple myeloma,” we postulate that DS may regulate HNSCC stem cells. Another line of evidence that “HNSCC contains cancer stem cells (CSCs) that have greater radioresistance and capacity to change replication dynamics in response to irradiation compared to non-clonogenic cells [35],” can help characterize the effects of radiotherapy on “CD44+/ALDH+” HNSCC stem cells derived from patients, providing screening for responsible patients, as “CD44+/ALDH+” HNSCC stem cells are of radioresistance. Hyaluronan (HA), an important glycosaminoglycan component of the extracellular matrix (ECM) and its major cell surface receptor, CD44, Nanog/Oct4/Sox2, have been suggested to be important cellular mediators influencing tumor progression and treatment resistance in head and neck cancer [36]. Personalized medicine-based approach can model the patterns of chemoresistance and tumor recurrence using ovarian cancer stem cell spheroids [37]. Gene set enrichment analysis and iPathway analysis identified signaling pathways with major implications to the pathobiology of cancer (e.g. TNFα, IFN, IL6/STAT, NF-κB) that are enriched in cisplatin-resistant ALDHhighCD44high cells when compared to control cells. Such pathway analysis establishes the relationship between CD44high/CD133high/CD117high cancer stem cells phenotypes and Cetuximab and Paclitaxel treatment responses in head and neck cancer cell lines [38].
Fourth, a new concept of “living with cancer subclones” or “co-habit with cancer subclones” [39] sounds odd and against the decades-long dominant trend of "targeted molecular destruction of cancer," however; its focus on modifying the tumor microenvironment [40] gains attention with clinically proven case reports. When the treatment benefits and the side effects sound odd, can the patients risk their lives for such an intervention? That is particularly promising given the effect of epigenetics [41], immunotherapy [42] and microbiome [43] on cáncer; all lead to a wait-and-watch approach to disease, due to the fact that we can monitor the relapse pathway [44].
CONCLUSION
In Summary, to achieve above benefits for a patient, we need to isolate the patient's CSCs (Which is the REAL CHALLENGE still!), and to determine the mechanism by which subclones of ALDHhighCD44high HNCSCs resist to drugs via single-cell transcriptome, as we show with breast cancer, with specified tissue-relevant tumor microenvironment [45]. Measuring survival of HNCSC CSC lines in presence of cisplatin (or other FDA approved drugs) in cellular models for cancer subclonal evolution [39] can help develop therapeutics to drive cancer cells to dormancy, which is a lifetime subclonal evolution process developmentally evolved with local environments to cancer. All the procedures are still ongoing and under way - They need long-term monitoring and confirmation if they indeed work.
ACKNOWLEDGEMENT
Support came from CHOC Children's Research Institute, CHOC Children's Foundation, CHOC-UCI Translation Research Awards (UCI – ICTS NIH program grants), and VALB Hospital. This work was supported in part by the National Institutes of Health (NCI, R01CA164509; NIEHS, R01ES021801-04S1).
DISCLOSURE
The authors declare no conflict of interest.
1. Li SC, Tachiki LM, Kabeer MH, Dethlefs BA, Anthony MJ, et al. (2014) Cancer genomic research at the crossroads: Realizing the changing genetic landscape as intratumoral spatial and temporal heterogeneity becomes a confounding factor. Cancer Cell Int 14: 115.
2. Li SC, Stucky A, Chen X, Kabeer MH, Loudon WG, et al. (2018) Single-cell transcriptomes reveal the mechanism for a breast cancer prognostic gene panel. Oncotarget 9: 33290-33301.
3. Turner KM, Sun Y, Ji P, Granberg KJ, Bernard B, et al. (2015) Genomically amplified Akt3 activates DNA repair pathway and promotes glioma progression. Proc Natl Acad Sci U S A 112: 3421-3426.
4. Allison KH, Sledge GW (2014) Heterogeneity and cancer. Oncology (Williston Park) 28: 772-778.
5. Harada T, Yamamoto E, Yamano HO, Aoki H, Matsushita HO, et al. (2018) Surface microstructures are associated with mutational intratumoral heterogeneity in colorectal tumors. J Gastroenterol 53: 1241-1252.
6. Meyer M, Reimand J, Lan X, Head R, Zhu X, et al. (2015) Single cell-derived clonal analysis of human glioblastoma links functional and genomic heterogeneity. Proc Natl Acad Sci U S A 112: 851-856.
7. Li SC, Vu LT, Ho HW, Yin HZ, Keschrumrus V, et al. (2012) Cancer stem cells from a rare form of glioblastoma multiforme involving the neurogenic ventricular wall. Cancer Cell Int 12: 41-54.
8. Economo MN, Viswanathan S, Tasic B, Bas E, Winnubst J, et al. (2018) Distinct descending motor cortex pathways and their roles in movement. Nature 563: 79-84.
9. Yang Z, Li C, Fan Z, Liu H, Zhang X, et al. (2017) Single-cell sequencing reveals variants in ARID1A, GPRC5A and MLL2 driving self-renewal of human bladder cancer stem cells. Eur Urol 71: 8-12.
10. Altmann DM (2018) A Nobel Prize-worthy pursuit: Cancer immunology and harnessing immunity to tumour neoantigens. Immunology 155: 283-284.
11. Wefers C, Schreibelt G, Massuger L, de Vries IJM, Torensma R (2018) Immune curbing of cancer stem cells by CTLs directed to NANOG. Front Immunol 9: 1412.
12. Abdollahpour-Alitappeh M, Razavi-Vakhshourpour S, Abolhassani M (2018) Development of a new anti-CD123 monoclonal antibody to target the human CD123 antigen as an AML cancer stem cell biomarker. Biotechnol Appl Biochem.
13. Li SC, Kabeer MH (2013) Designer immunotherapy specific for cancer. J Cell Sci Ther 4. Available at: http://dxdoiorg/104172/2157-70131000e116
14. Ugel S, Peranzoni E, Desantis G, Chioda M, Walter S, et al. (2012) Immune tolerance to tumor antigens occurs in a specialized environment of the spleen. Cell Rep 2: 628-639.
15. Schwarz RE, Hiserodt JC (1990) Effects of splenectomy on the development of tumor-specific immunity. J Surg Res 48: 448-453.
16. Lin CS, Bamodu OA, Kuo KT, Huang CM, Liu SC, et al. (2018) Investigation of ovatodiolide, a macrocyclic diterpenoid, as a potential inhibitor of oral cancer stem-like cells properties via the inhibition of the JAK2/STAT3/JARID1B signal circuit. Phytomedicine 46: 93-103.
17. McDermott SC, Rodriguez-Ramirez C, McDermott SP, Wicha MS, Nor JE (2018) FGFR signaling regulates resistance of head and neck cancer stem cells to cisplatin. Oncotarget 9: 25148-25165.
18. Kim J, Shin JH, Chen CH, Cruz L, Farnebo L, et al. (2017) Targeting aldehyde dehydrogenase activity in head and neck squamous cell carcinoma with a novel small molecule inhibitor. Oncotarget 8: 52345-52356.
19. Vu LT, Keschrumrus V, Zhang X, Zhong JF, Su Q, et al. (2015) Correction: Tissue elasticity regulated tumor gene expression: Implication for diagnostic biomarkers of primitive neuroectodermal tumor. PLoS One 10: e0128504.
20. Vu LT, Keschrumrus V, Zhang X, Zhong JF, Su Q, et al. (2015) Tissue elasticity regulated tumor gene expression: Implication for diagnostic biomarkers of primitive neuroectodermal tumor. PLoS One 10: e0120336.
21. Chen X, Wen Q, Stucky A, Zeng Y, Gao S, et al. (2018) Relapse pathway of glioblastoma revealed by single-cell molecular analysis. Carcinogenesis 39: 931-936.
22. Gao L, Zeng Y, Luo X, Chen Y, Kabeer MH, et al. (2018) Microfluidic enrichment of plasma cells improves treatment of multiple myeloma. Mol Oncol 12: 1004-1011.
23. Ohmura-Kakutani H, Akiyama K, Maishi N, Ohga N, Hida Y, et al. (2014) Identification of tumor endothelial cells with high aldehyde dehydrogenase activity and a highly angiogenic phenotype. PLoS One 9: e113910.
24. Shang Z, Xu Y, Liang W, Liang K, Hu X, et al. (2017) Isolation of cancer progenitor cells from cancer stem cells in gastric cancer. Mol Med Rep 15: 3637-3643.
25. Hida K, Maishi N, Akiyama K, Ohmura-Kakutani H, Torii C, et al. (2017) Tumor endothelial cells with high aldehyde dehydrogenase activity show drug resistance. Cancer Sci 108: 2195-2203.
26. Mackay A, Burford A, Molinari V, Jones DTW, Izquierdo E, et al. (2018) Molecular, pathological, radiological and immune profiling of non-brainstem pediatric high-grade glioma from the HERBY phase II randomized trial. Cancer Cell 33: 829-842.
27. Petrirena GJ, Masliah-Planchon J, Sala Q, Pourroy B, Frappaz D, et al. (2018) Recurrent extraneural sonic hedgehog medulloblastoma exhibiting sustained response to vismodegib and temozolomide monotherapies and inter-metastatic molecular heterogeneity at progression. Oncotarget 9: 10175-10183.
28. Ma X, Liu Y, Liu Y, Alexandrov LB, Edmonson MN, et al. (2018) Pan-cancer genome and transcriptome analyses of 1,699 paediatric leukaemias and solid tumours. Nature 555: 371-376.
29. Filbin MG, Tirosh I, Hovestadt V, Shaw ML, Escalante LE, et al. (2018) Developmental and oncogenic programs in H3K27M gliomas dissected by single-cell RNA-seq. Science 360: 331-335.
30. Zustovich F, Lombardi G, Pastorelli D, Farina P, Furini L, et al. (2010) Bevacizumab and glioblastomas, a single-centre experience: How disease history and characteristics may affect clinical outcome. Anticancer Res 30: 5213-5216.
31. Lopez GY, Van Ziffle J, Onodera C, Grenert JP, Yeh I, et al. (2018|) The genetic landscape of gliomas arising after therapeutic radiation. Acta Neuropathol.
32. Li SC, Han YP, Dethlefs BA, Loudon WG. (2010) Therapeutic window, a critical developmental stage for stem cell therapies. Curr Stem Cell Res Ther 5: 297-303.
33. Deng P, Wang J, Zhang X, Wu X, Ji N, et al. (2018) AFF4 promotes tumorigenesis and tumor-initiation capacity of head and neck squamous cell carcinoma cells by regulating SOX2. Carcinogenesis 39: 937-947.
34. Jin N, Zhu X, Cheng F, Zhang L (2018) Disulfiram/copper targets stem cell-like ALDH(+) population of multiple myeloma by inhibition of ALDH1A1 and Hedgehog pathway. J Cell Biochem 119: 6882-6893.
35. Reid P, Wilson P, Li Y, Marcu LG, Staudacher AH, et al. (2017) In vitro investigation of head and neck cancer stem cell proportions and their changes following X-ray irradiation as a function of HPV status. PLoS One 12: e0186186.
36. Bourguignon LYW, Earle C, Shiina M (2017) Activation of matrix hyaluronan-mediated CD44 signaling, epigenetic regulation and chemoresistance in head and neck cancer stem cells. Int J Mol Sci 18: E1849.
37. Raghavan S, Mehta P, Ward MR, Bregenzer ME, Fleck EMA, et al. (2017) Personalized medicine-based approach to model patterns of chemoresistance and tumor recurrence using ovarian cancer stem cell spheroids. Clin Cancer Res 23: 6934-6945.
38. Silva Galbiatti-Dias AL, Fernandes GMM, Castanhole-Nunes MMU, Hidalgo LF, Nascimento Filho CHV, et al. (2018) Relationship between CD44(high)/CD133(high)/CD117(high) cancer stem cells phenotype and cetuximab and paclitaxel treatment response in head and neck cancer cell lines. Am J Cancer Res 8: 1633-1641.
39. Li SC, Lee KL, Luo J (2012) Control dominating subclones for managing cancer progression and posttreatment recurrence by subclonal switchboard signal: Implication for new therapies. Stem Cells Dev 21: 503-506.
40. Li SC, Kabeer MH (2018) Spatiotemporal switching signals for cancer stem cell activation in pediatric origins of adulthood cancer: Towards a watch-and-wait lifetime strategy for cancer treatment. World J Stem Cells 10: 15-22.
41. Plass M, Solana J, Wolf FA, Ayoub S, Misios A, et al. (2018) Cell type atlas and lineage tree of a whole complex animal by single-cell transcriptomics. Science 360.
42. Shi T, Ma Y, Yu L, Jiang J, Shen S, et al. (2018) Cancer immunotherapy: A focus on the regulation of immune checkpoints. Int J Mol Sci 19.
43. Galluzzi L, Chan TA, Kroemer G, Wolchok JD, Lopez-Soto A (2018) The hallmarks of successful anticancer immunotherapy. Sci Transl Med 10.
44. Chen X, Wen Q, Stucky A, Zeng Y, Gao S, et al. (2018) Relapse pathway of glioblastoma revealed by single-cell molecular analysis. Carcinogenesis 39: 931-936.
45. Li SC, Vu LT, Luo JJ, Zhong JF, Li Z, et al. (2017) Tissue elasticity bridges cancer stem cells to the tumor microenvironment through microRNAs: Implications for a “watch-and-wait” approach to cancer. Curr Stem Cell Res Ther 12: 455-470.
QUICK LINKS
- SUBMIT MANUSCRIPT
- RECOMMEND THE JOURNAL
-
SUBSCRIBE FOR ALERTS
RELATED JOURNALS
- Journal of Renal Transplantation Science (ISSN:2640-0847)
- Journal of Cardiology and Diagnostics Research (ISSN:2639-4634)
- Journal of Clinical Trials and Research (ISSN:2637-7373)
- International Journal of AIDS (ISSN: 2644-3023)
- Journal of Forensic Research and Criminal Investigation (ISSN: 2640-0846)
- Ophthalmology Clinics and Research (ISSN:2638-115X)
- International Journal of Anaesthesia and Research (ISSN:2641-399X)